- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Pancreatic ductal adenocarcinoma (PDAC) is the deadliest cancer mainly owing to its proclivity to early metastasis and the lack of effective targeted therapeutic drugs. Hence,
understanding the molecular mechanisms underlying early invasion and metastasis by PDAC is imperative for improving patient outcomes. The present study identified that upregulation of TSPAN8
expression in PDAC facilitates metastasis in vivo and in vitro. We found SOX9 as a key transcriptional regulator of _TSPAN8_ expression in response to EGF stimulation. SOX9 modulation was
sufficient to positively regulate endogenous expression of TSPAN8, with concomitant in vitro phenotypic changes such as loss of cell–matrix adherence and increased invasion. Moreover,
increased SOX9 and TSPAN8 levels were shown to correlate in human pancreatic cancer specimens and downregulated in vitro by EGFR tyrosine kinase inhibitors. High expression of SOX9 and
TSPAN8 has been associated with tumor stage, poor prognosis and poor patient survival in PDAC. In conclusion, this study highlights the importance of the EGF-SOX9-TSPAN8 signaling cascade in
the control of PDAC invasion and implies that TSPAN8 may be a promising novel therapeutic target for the treatment of PDAC. SIMILAR CONTENT BEING VIEWED BY OTHERS S100A2 INDUCES
EPITHELIAL–MESENCHYMAL TRANSITION AND METASTASIS IN PANCREATIC CANCER BY COORDINATING TRANSFORMING GROWTH FACTOR Β SIGNALING IN SMAD4-DEPENDENT MANNER Article Open access 27 September 2023
MAP4K4 PROMOTES OVARIAN CANCER METASTASIS THROUGH DIMINISHING ADAM10-DEPENDENT N-CADHERIN CLEAVAGE Article Open access 15 March 2023 MACC1 PROMOTES PANCREATIC CANCER METASTASIS BY
INTERACTING WITH THE EMT REGULATOR SNAI1 Article Open access 04 November 2022 INTRODUCTION Pancreatic ductal adenocarcinoma (PDAC) is an extremely lethal cancer worldwide with limited
therapeutic options and a dismal prognosis. By 2030, PDAC is poised to become the second primary cause of cancer-related death [1, 2]. Metastasis is the leading cause of mortality and a
major driver behind the lethal nature of PDAC [3]. Clinically, most patients are diagnosed at a very late stage with metastatic dissemination, at which point the 5-year survival rate is only
3% [4]. Even in those patients who have had curative surgical resection with clear tumor margins (R0), 75% die of recurrence and metastasis within 5 years after operation [5]. The molecular
mechanisms involved in the metastatic cascade remain incompletely understood owing to the complexity of the process. Deep molecular insights into the metastatic process of PDAC are
imperative for the development of effective targeted treatment strategies. Tetraspanins are a superfamily of transmembrane proteins containing four highly hydrophobic transmembrane domains
(TMs) and N-terminal and C-terminal cytoplasmic tails [6]. In mammals, the tetraspanin family consists of 33 members, including clusters of differentiation-related protein 9 (CD9), CD37,
CD63, CD81 and tetraspanin-8 (TSPAN8; encoded by the gene _TSPAN8_). TSPAN8 has been implicated in many cellular functions because it forms tetraspanin-enriched microdomains (TEMs) with
different molecular chaperonins, such as cluster of differentiation (CD) proteins, including CD9, CD37, CD53, CD63 and CD81 [7], integrins, MHC class II antigens and T-cell receptors [8, 9].
Recent evidence suggests that TSPAN8 has an important role in tumor invasion and metastasis in multiple types of tumors, including PDAC [10], ovarian carcinoma [11], gastric adenocarcinoma
[12], colon adenocarcinoma [13], liver hepatocellular carcinoma [14], esophageal carcinoma [15], melanoma and glioma [16]. Margot Zöller et al. indicated that TSPAN8 can form a complex with
CD49c, CD9 and CD151 to promote cell migration by internalization [17] or can interact with integrin α6β4 [18]. TSPAN8 is a component of exosomes and increases sensitivity and specificity by
mediating the reprogramming of target cells [19,20,21]. TSPAN8 can mediate mesenchymal–epithelial transition by upregulating E-cadherin and downregulating Twist, P120-catenin and β-catenin
[22]. Our previous research revealed that the expression level of TSPAN8 is upregulated in breast cancer stem cells and correlates with chemotherapeutic resistance and poor prognosis [8].
Despite its well-established importance in tumor progression, metastasis and drug resistance, most previous studies have focused on how TSPAN8 interacts with other molecules and organizes
membrane networks as a ‘molecular facilitator' to achieve its biological functions [23]. However, why and how TSPAN8 expression is switched on during tumor progression are ill-defined.
Identifying factors influencing TSPAN8 expression, which enables acquisition of an invasive phenotype, is crucial to improving patient treatment and outcome. Sex-determining region Y-related
high-mobility group (HMG)-box (SOX) proteins are a family of transcription factors (TFs) highly expressed in multiple aggressive tumors [24]. Of the SOX family members, SOX9 is a vital TF
characterized by the existence of a SRY box, a 79-amino-acid motif that encodes a conserved HMG DNA-binding domain [25]. Initial studies show that SOX9 participates in the maintenance of
stemness in stem/progenitor cells and participates in relevant roles in organogenesis, such as progenitor differentiation, sex determination, oligodendrocyte development and neural crest
cell development [26,27,28,29]. Recent evidence indicates that SOX9 participates in cancer initiation and tumourigenicity via its regulation of initiating cells, which are functionally
linked with TGFβ/Smad, Notch and Wnt/β-catenin signaling activation [30,31,32]. Clinically, high SOX9 expression is correlated with metastasis, chemoresistance and poor prognosis in multiple
tumors [33,34,35,36,37,38,39]. In the pancreas, SOX9 is a main regulator of pancreatic progenitor cells and has a vital role in pancreatic endocrine and ductal cell differentiation during
pancreas development [39, 40]. Under chronic inflammation, the overexpression of SOX9 induced by epidermal growth factor (EGFR) signaling is indispensable for acinar to ductal metaplasia
(ADM) transdifferentiation [41] and for the KRAS-induced initiation of intraepithelial neoplasias [42, 43]. In this study, we demonstrated that upregulation of TSPAN8 expression in PDAC
promotes metastasis in vivo and in vitro. We identified SOX9 as a key transcriptional regulator of _TSPAN8_ overexpression in response to EGF stimulation. SOX9 depletion by short hairpin RNA
(shRNA) expression completely abrogated the enhanced expression of TSPAN8 at the protein and mRNA levels upon EGF treatment. Moreover, immunohistochemistry (IHC) staining analyses of human
PDAC specimens determined the correlation between EGFR, SOX9 and TSPAN8 expression. High-expression levels of SOX9 and TSPAN8 were associated with tumor stage, dismal prognosis and poor
survival in PDAC. These findings demonstrate the role of the EGF-SOX9-TSPAN8 signaling cascade during tumor metastasis and highlight the importance of TSPAN8 as a valuable therapeutic target
for PDAC treatment. RESULTS TSPAN8 IS HIGHLY EXPRESSED IN PDAC AND IS ASSOCIATED WITH PROGRESSION AND POOR PROGNOSIS The expression level of TSPAN8 in tumor tissues (TTs) and normal
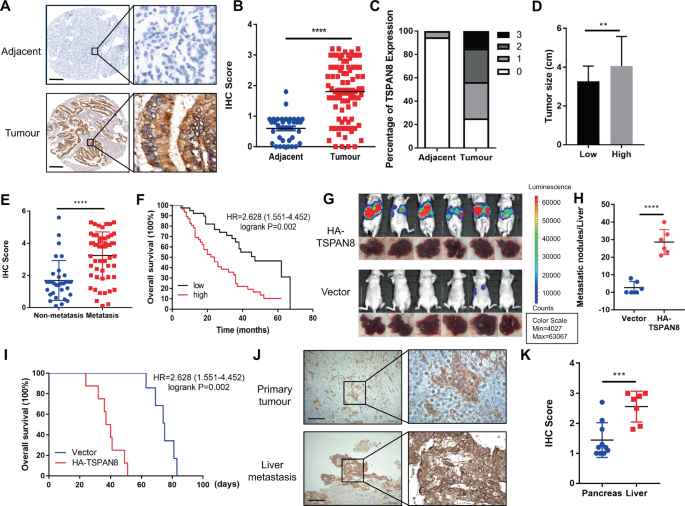
adjacent tissues (NATs) was determined by tissue microarray analysis, including 87 PDAC patients. IHC staining analysis showed that TSPAN8 was significantly highly expressed in TTs compared
with NATs (Fig. 1A–C, Supplementary Table 1). TSPAN8 expression levels were positively associated with tumor size (Fig. 1D). To further research the clinical significance of TSPAN8, TSPAN8
expression in PDAC patients with distant metastasis was compared with that in patients without distant metastasis. The results showed that TSPAN8 expression was significantly higher in PDAC
patients with metastasis (Fig. 1E). Furthermore, the expression levels of TSPAN8 were negatively correlated with the overall survival time of PDAC patients (_P_ = 0.002, Fig. 1F). In breast
cancer and liver hepatocellular carcinoma, high expressions of TSPAN8 were observed (Fig S1A, B). In addition, _TSPAN8_ mRNA data from the TCGA and GTEx databases were analyzed via the GEPIA
online tool (http://gepia.cancer-pku.cn). Consistently, we observed that the level of _TSPAN8_ mRNA was significantly enhanced in TTs compared with NATs. Similar phenomena were also
observed in colon adenocarcinoma, liver hepatocellular carcinoma, prostate carcinoma, rectal adenocarcinoma, stomach adenocarcinoma and esophageal carcinoma (Fig S1C). To further assess the
impact of TSPAN8 on PDAC metastasis, 106 SW1990 cells with or without HA-TSPAN8 expression were injected into athymic nude mice by intrasplenic injection to construct a liver metastasis
mouse model. SW1990 cells expressing HA-TSPAN8 showed a strong metastasis ability (Fig. 1G, H, Fig S1D, E) and shortened survival times (Fig. 1I). In contrast, TSPAN8 depletion reduced liver
metastasis and prolonged mouse survival time (data not shown). To determine the clinical relevance of TSPAN8 in PDAC metastasis, we performed IHC staining of primary tumor and liver
metastasis tissues from 17 PDAC patients. A significant increase in TSPAN8 expression in liver metastasis tissues compared with pancreatic primary TTs was observed (Fig. 1J, K). These
results suggest that TSPAN8 expression is positively correlated with tumor metastasis and poor prognosis in PDAC patients. TSPAN8 HAS A KEY ROLE IN PANCREATIC TUMOR CELL INVASION AND
MIGRATION To determine the biological roles of TSPAN8 in PDAC metastasis, we carried out an immunoblotting analysis of TSPAN8 expression in a panel of pancreatic cancer cell lines with
different metastatic potential (Fig. S2A). The results suggested that the expression level of TSPAN8 was significantly elevated in the malignant pancreatic cancer cell lines BxPC-3, AsPC-1
and SW1990 compared with the normal cell line HPDE6-C7 (Fig. 2A). In addition, the expression of TSPAN8 showed a positive relationship with the increasing metastatic ability of the cancer
cells. Consistent with the results of the protein expression levels, quantitative PCR analysis showed that _TSPAN8_ mRNA levels were significantly upregulated in PDAC cancer cells compared
with normal cells (Fig. 2B). Next, functional analysis was performed by cell migration and invasion assays. TSPAN8 depletion by _TSPAN8_-specific shRNA significantly reduced the invasion
ability of BxPC-3 and AsPC-1 cells (Fig. 2C–F). To further determine whether this effect resulted from TSPAN8 depletion specifically, BxPC-3 and AsPC-1 cells with TSPAN8 depletion were
administered shRNA-resistant TSPAN8 (rescue TSPAN8) (Fig S2B, C). As a result, the expression of rescue TSPAN8 significantly reversed the inhibitory effect of TSPAN8 depletion on cellular
invasion (Fig. 2C–F), suggesting that TSPAN8 has an important role in enhancing pancreatic cancer cell invasion. To further evaluate the metastatic functions of TSPAN8, TSPAN8 was
transiently overexpressed in HPDE6-C7 cells (Fig S2D). As shown in Fig S2E, F, the cell invasion ability was significantly enhanced by TSPAN8 overexpression. These results demonstrate that
TSPAN8 is important for the regulation of cellular invasion and tumor metastasis. SOX9 IS REQUIRED FOR EGF-INDUCED TSPAN8 UPREGULATION Epidermal growth factor (EGF)/EGFR signaling has been
implicated in many steps in the processes of tumor invasion and metastasis. In PDAC, EGFR is overexpressed in more than half of cases [44, 45]. To determine whether upregulation of TSPAN8
expression is a response to EGF, we evaluated the changes in TSPAN8 expression in BxPC-3 and SW1990 cells with or without EGF stimulation. QPCR analysis indicated that EGF stimulation
resulted in a dramatic increase in _TSPAN8_ mRNA in a time-dependent manner (Fig. 3A). In line with this, the protein expression levels of TSPAN8 were also upregulated upon EGF treatment
(Fig. 3B, Fig S3A, B). However, EGF-induced upregulation of _TSPAN8_ mRNA expression was suppressed by the EGFR tyrosine kinase inhibitors gefitinib (10 μM), erlotinib (10 μM) and AG1478 (10
μM) in BxPC-3 and SW1990 cells (Fig. 3C, Fig S3C). _TSPAN8_ mRNA expression is regulated by upstream promoters and TFs during tumor progression. To investigate the mechanism underlying
_TSPAN8_ upregulation, potential TFs of _TSPAN8_ were predicted via the JASPAR (http://jaspar.genereg.net) and AliBaba 2.1 (http://gene-regulation.com/pub/programs/alibaba2) online tools.
After combined analysis via JASPAR and AliBaba 2.1, 10 TFs were obtained. To assess the potential influence of predicted TFs on _TSPAN8_ mRNA expression, HPDE6-C7 cells with overexpression
of 10 TFs were generated. Overexpression of SOX9 and JUN significantly enhanced _TSPAN8_ mRNA expression, and SOX9 had the greatest influence (Fig. 3D). To further study the underlying
relationships, luciferase reporter assays were performed with Flag-SOX9 and Flag-JUN plasmids. As shown in Fig. 3E, SOX9, but not JUN, caused a significant change in the luciferase activity
of the _TSPAN8_ promoter-reporter construct, indicating that SOX9 might regulate _TSPAN8_ transcription. The mRNA and protein expression levels of SOX9 were further found to increase upon
EGF treatment but were reduced by gefitinib and erlotinib treatment in BxPC-3 cells (Fig. 3F, G), suggesting an EGF-dependent regulatory mechanism. To determine the key signaling molecule
that mediates SOX9 upregulation, a panel of inhibitors linked to EGFR signaling pathway kinases was used, and we found that SOX9 expression was significantly decreased by treatment with the
ERK inhibitor GDC-0994 but not the AKT inhibitor compound 26 or the PKCα inhibitor GO6983 in BxPC-3 cells (Fig. 3F, G), consistent with a previous report [46, 47]. To study the
transcriptional regulation of SOX9 to _TSPAN8_, SOX9 depletion by _SOX9_-specific shRNA was performed in BxPC-3 cells (Fig S3D, E). As expected, the results of qPCR and immunoblot analyses
showed that the enhanced expression of TSPAN8 at the mRNA and protein levels upon EGF treatment was inhibited by _SOX9_-specific shRNA in BxPC-3 cells (Fig. 3H, I). In addition, HCC827
cells, which have constitutively active EGFR, were treated with or without shSOX9, and the results showed that TSPAN8 was high even without EGF stimulation, but shSOX9 reversed this increase
(Fig. 3H, I). To further confirm the EGF-induced upregulation of TSPAN8 expression, BxPC-3 and SW1990 cells were treated with gefitinib, erlotinib or AG1478 under EGF stimulation, and
immunoblotting was performed. The results showed that gefitinib, erlotinib and AG1478 inhibited the phosphorylation of EGFR and ERK, which reduced SOX9 expression and reversed the increase
in TSPAN8 expression caused by EGF stimulation (Fig. 3J, Fig S3F). These data indicate that SOX9 is required for the upregulation of _TSPAN8_ expression mediated by EGF. SOX9 REGULATES
_TSPAN8_ MRNA EXPRESSION BY DIRECTLY BINDING TO ITS PROMOTER Since SOX9 is known as a TF [48] and our results determined that SOX9 is responsible for the upregulation of _TSPAN8_ mRNA
expression upon EGF/EGFR activation, we wondered whether SOX9 could transcriptionally regulate _TSPAN8_ expression in PDAC. To further investigate this result, chromatin IP sequencing was
performed with an anti-Flag antibody in HDPE6-C7 cells stably overexpressing Flag-SOX9, and 10625 different peaks mapping to 1077 genes were obtained (Fig S4A). To examine the functions of
these genes, we performed GO term enrichment analysis. The GO terms response to stimulus, cell migration, motility and growth were enriched with these genes, which include EGFR signaling
pathway-related genes (Fig S4B, Supplementary Table 2). All enriched GO terms with _P_ < 0.05 are summarized in Supplementary Table 3. KEGG pathway enrichment analysis showed that the
regulation of the pathways of the actin cytoskeleton and PI3K-AKT signaling were enriched (Fig S4C). Meanwhile, four promoter fragments of _TSPAN8_ were identified. We discovered that
Flag-SOX9 dramatically enhanced luciferase intensity at residues 758–963 and 1330–1543 upstream of the transcriptional start site (TSS) in the promoter region of _TSPAN8_ (Fig. 4A). By
comparing the binding motifs of SOX9 and the promoter sequence of _TSPAN8_, we identified two binding sites, namely, 1340–1353 and 815–821, upstream of the TSS of _TSPAN8_, which is
consistent with previous results [25] (Fig. 4B, Fig S4D). Thereafter, we found that a SOX9 construct with one of the binding sites mutated had a less powerful effect on increasing the
expression of _TSPAN8_ than the wild-type SOX9 construct. When the two binding sites were mutated simultaneously, SOX9 was not able to induce any activity of the _TSPAN8_ promoter (Fig. 4C).
ChIP-qPCR analysis confirmed the binding of SOX9 to _TSPAN8_ at specific sites (Fig. 4D). Consistent with this finding, electromobility shift assays (EMSAs) showed that SOX9 binds two sites
in the promoter of _TSPAN8_ (Fig. 4E). Collectively, these data suggest that SOX9 promotes the transcription of _TSPAN8_ by binding to two sites in the promoter of _TSPAN8_. TSPAN8 AND SOX9
EXPRESSION LEVELS ARE CORRELATED, AND HIGH-EXPRESSION LEVELS OF BOTH ARE RELATED TO POOR PROGNOSIS To identify the correlation between the expression levels of TSPAN8, SOX9 and EGFR in
PDAC, we performed IHC analysis of an additional cohort of 40 human pancreatic cancer specimens. Pearson correlation analysis depicted a positive association between the expression levels of
TSPAN8 and SOX9, as well as between TSPAN8 and EGFR (Fig. 5A, B). Finally, we analyzed the correlation of _SOX9_ and _TSPAN8_ in the TCGA data set using the cBioPortal online tool
(http://www.cbioportal.org). The results showed that the mRNA expression of _TSPAN8_ was significantly correlated with _SOX9_ in multiple cancers (Fig. 5C, Fig S5A–C). Furthermore, through
Kaplan–Meier Plotter (https://kmplot.com/analysis), we found that high-expression levels of SOX9 or TSPAN8 were correlated with poor prognosis in PDAC patients (Fig. 5D). Similar results
have also been observed in other cancers, such as liver hepatocellular carcinoma and breast cancer. Collectively, these results strongly suggest that EGF-SOX9 signaling promotes PDAC
progression by enhancing _TSPAN8_ transcription. DISCUSSION Emerging evidence supports that TSPAN8 has an important role in tumor progression and metastasis [11, 49]. Our previous study
demonstrated that TSPAN8 plays a key role in the regulation of breast cancer cell stemness via activation of sonic hedgehog signaling [8]. Here, we demonstrated that TSPAN8 overexpression
promoted PDAC cells invasion and migration. Clinically, TSPAN8 expression levels were positively linked with tumor stage, size and axillary node metastasis and inversely correlated with the
overall survival time in PDAC patients. Moreover, TSPAN8 overexpression markedly enhanced liver metastasis in vivo and shortened mouse survival time. Despite the central role of TSPAN8 in
critical processes for tumor progression, the underlying molecular mechanisms of TSPAN8 expression upregulation remain poorly understood [50]. In our attempts to investigate the mechanism
underlying TSPAN8 overexpression in PDAC cells, we found that SOX9 is a critical regulator of _TSPAN8_ transcription. The developmental regulator SOX9 is widely linked to cancer cell
proliferation, progression, survival and evasion of senescence in different cancers [39, 51, 52]. In PDAC, SOX9 has oncogenic roles to promote progression and metastasis [41, 53]. In
addition, the expression of SOX9 is induced by EGF/EGFR signaling activation [53]. Our results showed that SOX9 expression was upregulated through the EGFR-ERK signaling axis. Given that
SOX9 was identified as a potential TF of _TSPAN8_ by sequence analysis, we speculated that TSPAN8 overexpression might be caused by SOX9 transcriptional regulation. Immunoblotting and qPCR
analyses showed that SOX9 depletion by shRNA completely abrogated the enhanced expression of TSPAN8 induced upon EGF treatment at the protein and mRNA levels. A further mechanistic study
indicated that SOX9 bound to two sites of the _TSPAN8_ promoter to promote its transcription. Consequently, TSPAN8 expression promoted PDAC invasion and metastasis (Fig. 6). Our analyses of
human PDAC specimens depicted a positive correlation among the expression levels of TSPAN8, EGFR and SOX9. Of note, TSPAN8 and SOX9 expression levels were related to poor prognosis in
patients with PDAC and other tumors. Taken together, our results illustrate a novel regulatory role of EGF-ERK-SOX9-TSPAN8 signaling in cancer cell migration and invasion. Our determination
of the mechanism by which SOX9 is upregulated under EGF activation and promotes TSPAN8 transcription provides insight for determining how cell motility is regulated in response to
environmental stimuli. The EGF/EGFR signaling pathway is frequently deregulated in PDAC. To date, only one targeted drug, erlotinib, which is an orally administered EGFR tyrosine kinase
inhibitor, has been approved for the treatment of PDAC. However, its effectiveness has long been questioned due to its low response rate and overall survival rate [54]. This maybe due to the
enrichment of TSPAN8 in exosomes. The function of exosomes in tumor metastasis has been widely reported [55]. TSPAN8, as a tetraspanin, form the complexes with other tetraspanin proteins as
constitutive components of exosomes to active the signaling cascades in target cells [17, 19,20,21, 56]. This counteracts the positive response of erlotinib, decreases the overall survival
rate and promotes tumor metastasis in PDAC finally. The establishment of the critical role of the EGF-ERK-SOX9-TSPAN8 signaling cascade in promoting tumor metastasis makes TSPAN8 a novel
therapeutic target for the treatment of PDAC. MATERIALS AND METHODS CELL CULTURE Capan-2, PANC-1, HEK293T and HCC827 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium
(DMEM, HyClone, Illinois, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, New York, USA), 100 U/ml penicillin and 100 mg/ml streptomycin (HyClone, Illinois, USA) at 37 °C with 5%
CO2. HPDE6-C7, SW1990, AsPC-1 and BxPC-3 cells were cultured in Roswell Park Memorial Institute-1640 medium (HyClone, Illinois, USA) supplemented with 10% FBS, 100 U/ml penicillin and 100
mg/ml streptomycin at 37°C with 5% CO2. EGF treatment was administered at a final concentration of 100 ng/ml. All of the cell lines used in this study were purchased from ATCC and routinely
tested for mycoplasma contamination every 2 weeks. DNA CONSTRUCTS AND MUTAGENESIS For the generation of cell lines with gene depletion or overexpression, the viral skeleton plasmid
pLKO.1-Hygro and the EGFP-tagged HA/3xFlag-PGK-Puro vector were transfected into the indicated cell lines to silence and overexpress TSPAN8, respectively. After antibiotic selection, the
knockdown efficiency and overexpression efficiency were assessed by immunoblotting. Cells with depleted endogenous TSPAN8 and reconstituted cells with stable expression of shRNA-resistant
TSPAN8 (rescue TSPAN8) were utilized for immunoblotting analysis, qPCR assays, cell invasion analysis and tumor xenograft experiments as indicated in our study. pGL3 vectors containing
_TSPAN8_ WT and mutant promoters were constructed for the luciferase reporter gene assay. The shRNA and PCR primer oligonucleotide sequences used in our research are shown in Supplementary
Table 4. MATERIALS Antibodies that recognize TSPAN8 (ab70007, 1:1000), HA (#3724, 1:5000), EGFR (ab52894, 1:8000) and phospho-EFGR Y1068 (ab32430, 1:8000) were purchased from Abcam
(Cambridge, UK). Anti-SOX9 (#82630, 1:1000), β-actin (#4970, 1:5000), phospho-p44/42 MAPK (Erk 1/2) T202/Y204, rabbit horseradish peroxidase (HRP)-linked (#7074, 1:5000) and mouse HRP-linked
(#7076, 1:5000) antibodies were purchased from Cell Signaling Technology (Massachusetts, USA). Anti-ERK1/2 (16443–1-AP, 1:1000) and phospho-AKT S473 (66444–1-lg, 1:1000) were purchased from
Proteintech Group, Inc (Chicago, USA). Anti-phospho-PKCα S657 (sc-377565, 1:100) was purchased from Santa Cruz Biotechnology (Texas, USA). Recombinant human EGF (rhEGF, PHG0311L) was
purchased from Gibco (New York, USA). D-luciferin sodium salt (P1043) was purchased from Promega (Wisconsin, USA). Gefitinib (HY-50895), Erlotinib (HY-50896), AG1478 (HY-13524), Compound 26
(HY-18296), GDC-0994 (HY-15947) and GO6983 (HY-13689) were purchased from MedChemExpress (MCE) Company (Shanghai, China). TRANSFECTION Cells were plated at a density of 1 × 106 per 10 cm
dish 24 hours before transfection. Transfection was performed as previously described [8]. IMMUNOBLOTTING ANALYSIS Proteins were extracted from cultured cells using RIPA buffer (Beyotime,
Shanghai, China) at 4°C followed by immunoblotting with the corresponding antibodies in the presence of protease inhibitor cocktail and phosphatase inhibitor cocktail. The protein
concentration was determined using a BCA Protein Assay Kit (Beyotime, Shanghai, China). Proteins from cell lysates were separated by SDS-PAGE, transferred onto polyvinylidene difluoride
membranes (Millipore Corporation, Massachusetts, USA) and probed with the indicated primary antibodies and HRP-conjugated secondary antibodies. GENE EXPRESSION ANALYSIS We isolated total RNA
from cells using a Tissue RNA Kit (Biomiga Corporation, San Diego, USA) following the manufacturer’s instructions. We synthesized cDNA from 500 ng total RNA using the PrimerScript RT
Reagent Kit (Takara Corporation, Dalian, China) and quantified mRNA levels by qPCR using the SYBR Premix Ex Taq Kit (Takara Corporation, Dalian, China). We ran samples in technical
triplicates, and the calculated mRNA levels of the genes of interest were normalized to GAPDH mRNA levels in the same samples using the 2−∆∆CT method. The primers utilized in our
investigation are listed in Supplementary Table 5. CELL MIGRATION AND INVASION ASSAYS Cells were seeded in six-well plates, scratched with 10 µl pipet tips and cultured in 1× DMEM solution
without serum. Then, the cells were washed three times with phosphate-buffered saline (PBS) and photographed by microscopy (Leica, Wetzlar, Germany) at 0 hours, 6 hours and 12 hours. The
migration distance was measured by Image-Pro Plus software. Cells were seeded in 24-well invasion chambers (BD Biosciences, New Jersey, USA) with a Matrigel-coated film insert (8 mm pore).
The mixed solution was diluted to generate a 1× DMEM solution containing 10% serum. Two days later, cells on the bottom surface of the filter were subjected to staining with crystal violet
for 15 min and then washed three times with PBS, and the cell number was counted under a microscope (Leica, Wetzlar, Germany). HUMAN TISSUE SPECIMENS AND IMMUNOHISTOCHEMICAL ANALYSIS The
experiment with human tissues was authorized by the Human Ethics Committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China). All subjects
provided written informed consent. Patients with radiotherapy or chemotherapy treatment before surgery were excluded. Survival time was calculated from the date of surgery to the date of
death or last follow-up. Tumor-node-metastasis staging was performed according to American Joint Committee on Cancer (AJCC)/Union for International Cancer Control (UICC) standards. IHC
analysis was performed as previously described [8]. The pathological types of paraffin-embedded slides were checked again by HE staining before IHC analysis for TSPAN8, EGFR and SOX9. A
rabbit polyclonal anti-TSPAN8 antibody (1:100, Abcam, ab7007), rabbit monoclonal anti-EGFR antibody (1:100, Abcam, ab52894) and rabbit monoclonal anti-SOX9 antibody (1:400, CST, #82630) were
used. A DAB Substrate Kit (Zsbio Commerce Store) was used according to the manufacturer’s instructions. The scores for staining frequency (0 = 0%, 1 = 1%, 2 = 2–10%, 3 = 11–30%, 4 = 31–70%
and 5 = 71–100%) and intensity (0 = negative, 1 = week, 2 = moderate and 3 = strong staining) were used. A DAB Substrate Kit (Zsbio Commerce Store) was used according to the manufacturer =
=negative, 1 were used. A DAB Substrate Kit (Zsbio Commerce Store) was used according to the manufacturer = week, 2 were used. A DAB Substrate Kit (Zsbio Commerce Store) was used according
to the manufacturer = moderate and 3 were used. A DAB Substrate Kit (Zsbio Commerce Store) was used according to the manufacturer = strong staining) were summed to obtain an overall staining
score (OSS). An OSS of 0–2 was deemed low, 3–5 was deemed moderate and 6–8 was deemed high. The results were scored by two pathologists blinded to the clinicopathologic data.
ELECTROMOBILITY SHIFT ASSAY Nuclear extracts from 107 HeLa cells transfected with Flag-SOX9 plasmid were prepared, and SOX9 was purified by immunoprecipitation (IP) using a Flag antibody for
EMSA as previously described [8, 57]. DNA probes were purchased from Sangon Biotech (Shanghai, China). EMSAs were performed by incubating samples with purified Flag-SOX9 protein in binding
buffer with the Gel Shift Assay Systems kit (Promega, Wisconsin, USA) according to the manufacturer’s protocol. Following incubation, samples were loaded onto a 4.5% native acrylamide gel
and electrophoresed for 30 min at 130 V. Gels were scanned using a ChemiDoc imager (Bio-Rad Laboratories, California, USA.) XENOGRAFT TUMOR STUDIES The animal experiment was performed as
previously described [58]. Twenty-four female Balb/c nude mice (5 weeks old) were divided into two groups (six mice per group): a group receiving SW1990 cells with overexpression of TSPAN8
and a group receiving SW1990 cells without overexpression of TSPAN8. A small left abdominal flank incision was made, the spleen was exteriorized, and the prepared cells (1 × 106 cells/50
µl/mouse) were injected into the spleen with a 30-gauge needle. To prevent tumor cell leakage and bleeding, a cotton swab was held over the site of injection for 5 min. The blood vessels of
the injected spleen were ligated, and the injected spleen was removed. The wound was sutured with 6–0 black silk. Six weeks later, all of the mice were sacrificed and necropsied for
observation of visible metastatic lesions in the liver. All animal experiments were approved by the animal care and use committee of Shanghai Renji Hospital, Shanghai Jiaotong University
School of Medicine. STATISTICAL ANALYSIS Differences between groups were analyzed using Student’s _t_ test, chi-square tests or Fisher’s exact test with GraphPad Prism 7.0 and SPSS 17.0
software. Pearson’s test was applied to determine the correlation between clinicopathological parameters and protein expression. Data are presented as the mean ± SD. Differences at _P_ <
0.05 were considered statistically significant. DATA AVAILABILITY All relevant data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES * Pishvaian MJ, Blais EM, Brody JR, Lyons E, DeArbeloa P, Hendifar A, et al. Overall survival in patients with pancreatic cancer reveiving matched therapies following molecular
profiling: a restrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020;21:508–18. Article CAS PubMed PubMed Central Google Scholar * Cave DD, Di Guida M, Costa
V, Sevillano M, Ferrante L, Heeschen C, et al. TGF-beta1 secreted by pancreatic stellate cells promotes stemness and tumourigenicity in pancreatic cancer cells through L1CAM downregulation.
Oncogene. 2020;39:4271–85. Article CAS PubMed PubMed Central Google Scholar * Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic
cancer with nab-paclitaxel plus gemcitabine. N. Engl J Med. 2013;369:1691–703. Article CAS Google Scholar * Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer.
Lancet. 2011;378:607–20. Article PubMed PubMed Central Google Scholar * Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. Article PubMed Google
Scholar * Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci. 2014;127:3641–8. CAS PubMed Google Scholar * Szala S, Kasai Y, Steplewski Z, Rodech U,
Koprowski H, Linnenbach AJ. Molecular cloning of cDNA for the human tumor-associated antigen CO-029 and identification of realted transmembrane antigens. Proc Nati Acad Sci USA.
1990;87:6833–7. Article CAS Google Scholar * Zhu R, Gires O, Zhu L, Liu J, Li J, Yang H, et al. TSPAN8 Promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat
commun. 2019;10:2863. Article PubMed PubMed Central CAS Google Scholar * Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel
type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. Article CAS PubMed Google Scholar * Wang H, Rana S, Giese N, Buchler MW, Zoller M. Tsapn8, CD44v6 and alpha6beta4
are biomarkers of migrating pancreatic cancer initiating cells. Int J Cancer. 2013;133:416–26. Article CAS PubMed Google Scholar * Park CS, Kim TK, Kim HG, Kim YJ, Jeoung MH, Lee WR, et
al. Therapeutic targeting of tetraspanin 8 in epithelial ovarian cancer invasion and metastasis. Oncogene. 2016;35:4540–8. Article CAS PubMed Google Scholar * Anami K, Que N, Noguchi T,
Sakanoto N, Sentani K, Hayashi T, et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer.
2016;19:370–80. Article CAS PubMed Google Scholar * Kin TK, Park CS, Jeoung MH, Lee WR, Go NK, Choi JR, et al. Generation of a human antibody that inhibits TSPAN8 mediated invasion of
metastatic colorectal cancer cells. Biochem Biophys Res Commun. 2015;468:774–80. Article CAS Google Scholar * Fang T, Lin J, Wang Y, Chen G, Huang J, Chen J, et al. Tetraspanin 8 promotes
hepatocellular carcinoma metastasis by increasing ADAM12m expression. Oncotarget. 2016;7:40630–43. Article PubMed PubMed Central Google Scholar * Yue GG, Lee JK, Li L, Chan KM, Wong EC,
Chan JY, et al. Andrographis paniculata elicits anti-invasion activities by suppressing TM4SF3 gene expression and by anoikis-sensitization in esophageal cancer cells. Am J Cancer Res.
2015;5:3570–8. CAS PubMed PubMed Central Google Scholar * EI Kharbili M, Agaesse G, Barbollat-Boutrand L, Pommier RM, de la Fouchardiere A, Larue L, et al. Tspan8-beta-catenin positive
feedback loop promotes melanoma invasion. Oncogene. 2019;38:3781–93. Article CAS Google Scholar * Rana S, Claas C, Cretz CC, Nazarenko I, Zoeller M. Activation-induced internalization
differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int J Biochem Cell Biol. 2011;43(1):106–9. Article CAS PubMed Google Scholar * Gesierich S, Paret C,
Hildebrand D, Weitz J, Zgraggen K, Schmitz-Winnenthal FH, et al. Colocalization of the tetraspanins, CO-029 and CD151, with integrins in human pancreatic adenocarcinoma: impact on cell
motility. Clin Cancer Res. 2005;11(8):2840–52. Article CAS PubMed Google Scholar * Yue SJ, Mu W, Erb U, Zöller M. The tetraspanins CD151 and Tspan8 are essential exosome components for
the crosstalk between cancer initiating cells and their surrounding. Oncotarget. 2015;6(4):2366–84. Article PubMed Google Scholar * Mu W, Provaznik J, Hackert T, Zöller M. Tspan8-tumor
extracellular vesicle-induced endothelial cell and fibroblast remodeling relies on the target cell-selective response. Cells. 2020;9(2):319. Article CAS PubMed Central Google Scholar *
Kyuno D, Zhao K, Bauer N, Ryschich E, Zöller M. Therapeutic targeting cancer-initiating cell markers by exosome miRNA: Efficacy and functional consequences exemplified for claudin7 and
EpCAM. Transl Oncol. 2019;12(2):191–9. Article PubMed Google Scholar * Voglstaetter M, Thomsen AR, Nouvel J, Koch A, Jank P, Navarro EG, et al. Tspan8 is expressed in breast cancer and
regulates E-cadherin/catenin signalling and metastasis accompanied by increased circulating extracellular vesicles. J Pathol. 2019;248(4):421–37. Article CAS PubMed PubMed Central Google
Scholar * Greco C, Bralet MP, Ailane N, Dubart-Kupperschmitt A, Rubinstein E, Le Naour F, et al. E-cadherin/p120-catenin and tetraspanin CO-029 cooperate for cell motility control in human
colon carcinoma. Cancer Res. 2010;70:7674–83. Article CAS PubMed Google Scholar * Luanpitpong S, Li J, Manke A, Brundage K, Ellis E, McLaughlin SL, et al. SLUG is required for SOX9
stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene. 2016;35:2824–33. Article CAS PubMed Google Scholar * Shi Z, Chiang CI, Labhart
P, Zhao Y, Yang J, Mistretta TA, et al. Context-specific role of SOX9 in NF-Y mediated gene regulation in colorectal cancer cells. Nucleic Acids Res. 2015;43:6257–69. Article CAS PubMed
PubMed Central Google Scholar * Pritchett J, Athwal V, Roberts N, Hanley NA, Hanley KP. Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol Med.
2011;17:166–74. Article CAS PubMed Google Scholar * Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–93. Article
CAS PubMed Google Scholar * Kawaguchi Y. Sox9 and programming of liver and pancreatic progenitors. J Clin Invest. 2013;123:1881–6. Article CAS PubMed PubMed Central Google Scholar *
Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–28. Article CAS PubMed
PubMed Central Google Scholar * Liu H, Liu Z, Jiang B, Peng R, Ma Z, Lu J. SOX9 overexpression promotes glioma metastasis via Wnt/beta-catenin signaling. Cell Biochem Biophys.
2015;73:205–12. Article CAS PubMed Google Scholar * Hu B, Wang J, Jin X. MicroRNA-138 suppresses cell proliferation and invasion of renal cell carcinoma by directly targeting SOX9. Oncol
lett. 2017;14:7583–8. PubMed PubMed Central Google Scholar * Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Eagon EN, et al. Distinct EMT programs control normal mammary stem cells and
tumour-initiating cells. Nature. 2015;525:256–60. Article CAS PubMed PubMed Central Google Scholar * Tang L, Jin J, Xu K, Wang X, Tang J, Guan X. SOX9 interacts with FOXC1 to activate
MYC and regulate CDK7 inhibitor sensitivity in triple-negative breast cancer. Oncogenesis. 2020;9:47. Article CAS PubMed PubMed Central Google Scholar * Higashihara T, Yoshitomi H,
Nakata Y, Kagawa S, Takano S, Shimizu H, et al. Sex determining region Y box 9 induces chemoresistance in pancreatic cancer cells by induction of putative cancer stem cell characteristics
and its high expression predicts poor prognosis. Pancreas. 2017;46:1296–304. Article CAS PubMed Google Scholar * Larsimont JC, Youssef KH, Sánchez-Danés A, Sukumaran V, Defrance M,
Delatte B, et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell. 2015;17:60–73. Article CAS PubMed Google Scholar *
Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell.
2012;21:601–13. Article CAS PubMed PubMed Central Google Scholar * Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. Prognostic significance of
cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med (Maywood). 2011;236:145–55. Article CAS Google Scholar * Yan S, Shan X, Chen K, Liu Y, Yu G, Chen
Q, et al. LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene. 2018;679:138–49. Article CAS PubMed Google Scholar *
Grimont A, Pinho AV, Cowley MJ, Augereau C, Mawson A, Giry-Laterriere M, et al. SOX9 regulates ERBB signalling in pancreatic cancer development. Gut. 2015;64:1790–9. Article CAS PubMed
Google Scholar * Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, et al. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell
differentiation. Development. 2012;139:2488–99. Article CAS PubMed PubMed Central Google Scholar * Hessmann E, Zhang JS, Chen MN, Hasselluhn M, Liou GY, Storz P, et al. NFATc4 regulates
Sox9 gene expression in acinar cell plasticity and pancreatic cancer initiation. Stem Cells Int. 2016;2016:5272498. Article PubMed CAS Google Scholar * Wang LD, Yang HB, Zamperone A,
Diolati D, Palmbos PL, Abel EV, et al. ATDC is required for the initiation of KRAS-induced pancreatic tumorigenesis. Genes Dev. 2019;33:641–55. Article CAS PubMed PubMed Central Google
Scholar * Kopp JL, Figura GV, Mayes E, Liu FF, Dubois CL, Morris JP, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of
pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–50. Article CAS PubMed PubMed Central Google Scholar * Park SJ, Gu MJ, Lee DS, Yun SS, Kim HJ, Chol JH. EGFR expression in
pancreatic intraepithelial neoplasia and ductal adenocarcinoma. Int J Exp Pathol. 2015;8:8298–304. Google Scholar * Nedaeinia R, Avan A, Manian M, Salehi R, Ghayour-Mobarhan M. EGFR as a
potential target for the treatment of pancreatic cancer: dilemma and controversies. Curr Drug Targets. 2014;15:1293–301. Article CAS PubMed Google Scholar * Ling SZ, Chang XF, Schultz L,
Lee KT, Chaux A, Marchionni L, et al. An EGFR-ERK-SOX9 signaling cascade links urothelial development and regeneration to cancer. Cancer Res. 2011;71(11):3812–21. Article CAS PubMed
PubMed Central Google Scholar * Kaushik G, Seshacharyulu P, Rauth S, Nallasamy P, Rachagani S, Nimmakayala RK, et al. Selective inhibition of stemness through EGFR/FOXA2/SOX9 axis reduce
pancreatic cancer metastasis. Oncogene. 2021;40(4):848–62. Article CAS PubMed Google Scholar * Shi J, Guo J, Li X. Role of LASP‐1, a novel SOX9 transcriptional target, in the progression
of lung cancer. Int J Oncol. 2018;52:179–88. CAS PubMed Google Scholar * Giampieri R, Piva F, Occhipinti G, Bittoni A, Righetti A, Pagliaretta S, et al. Clinical impact of different
exosomes’ protein expression in pancreatic ductal carcinoma patients treated with standard first line palliative chemotherapy. PLoS ONE. 2019;14:e309
https://doi.org/10.1371/journal.pone.0215990. Article Google Scholar * Agaesse G, Barbollat-Boutrand L, El Kharbili M, Berthier-Vergnes O, Masse I. p53 targets TSPAN8 to prevent invasion
in melanoma cells. Oncogenesis. 2017;6:e309 https://doi.org/10.1038/oncsis.2017.11. Article CAS PubMed PubMed Central Google Scholar * Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A,
Zhang Y, et al. Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem cells. 2013;31:1454–66. Article CAS PubMed PubMed Central Google
Scholar * Zhou H, Qin Y, Ji S, Ling J, Fu J, Zhuang Z, et al. SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene. 2018;37:912–23.
Article CAS PubMed Google Scholar * Chen MN, Singh G, Koenig A, Liou GY, Storz P, Zhang JS, et al. NFATc1 Links EGFR signaling to induction of Sox9 transcription and acinar-ductal
transdifferentiation in the pancreas. Gastroenterology. 2015;148:1024–34. Article CAS PubMed Google Scholar * Blasco MT, Navas C, Martín-Serrano G, Graña-Castro O, Lechuga CG., Martín
DíazL, et al. Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-RAF. Cancer Cell. 2019;35:573–87. Article CAS PubMed Google Scholar
* Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. Article CAS PubMed PubMed Central Google Scholar * Mu W, Wang
Z. Ping-pong-tumour and host in pancreatic cancer progression. Front Oncol. 2019;9:1359. Article PubMed PubMed Central Google Scholar * Si JY, Yu XY, Zhang YJ, James WD. Myc interacts
with Max and Miz1 to repress C/EBP delta promoter activity and gene express. Mol Cancer. 2010;9:92. Article PubMed PubMed Central CAS Google Scholar * Chen T, Li JJ, Xu MD, Zhao Q, Hou
YY, Yao LQ, et al. PKCε phosphorylates MIIP and promotes colorectal cancer metastasis through inhibiton of Rel A deacetylation. Nat Coummun. 2017;8:939. Article CAS Google Scholar
Download references ACKNOWLEDGEMENTS This work was supported by the National Natural Science Funds (grant number 82073269 and 81772802), Shanghai Science and Technology Innovation Action
Plan (grant number 20XD1402800) and Clinical Research Plan of SHDC (grant number SHDC2020CR2065B) to H.X.W., the Construction Project of Shanghai Key Laboratory of Molecular Imaging (grant
number 18DZ2260400) and Shanghai Municipal Education Commission (Class II Plateau Disciplinary Construction Program of Medical Technology of SUMHS, 2018–2020) to J.J.L. and the Huzhou
Natural Science Funds (grant number 2017YZ11) to Z.H.L. AUTHOR INFORMATION Author notes * These authors contributed equally: Junjian Li, Xiaoliang Chen, Liqun Zhu, Zhenghong Lao. AUTHORS AND
AFFILIATIONS * Shanghai Key Laboratory of Molecular Imaging, Shanghai University of Medicine and Health Sciences, Shanghai, China Junjian Li * State Key Laboratory of Oncogenes and Related
Genes, Department of Oncology, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China Junjian Li, Tianhao Zhou, Weiyu Ge, Jingxuan Xu, Yuan Cao,
Shaoqian Du & Hongxia Wang * The Center for Chronic Disease Control and Prevention, Shenzhen Guangming District Centers for Disease Control and Prevention, Shenzhen, China Xiaoliang Chen
* Department of Oncology, Liyang People’s Hospital, Liyang, China Liqun Zhu * Department of Oncology, Deqing People’s Hospital, Huzhou, China Zhenghong Lao * Pathology Center, Shanghai
General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China Lijuan Zang * Department of Medical Oncology, Shanghai Jiaotong University Affiliated Sixth People’s
Hospital East Campus, Shanghai, China Mengyi Jiang * Shanghai Experimental School, Shanghai, China Yue Yu * Translational Medicine Center, Shanghai General Hospital, Shanghai Jiao Tong
University School of Medicine, Shanghai, China Guangjian Fan Authors * Junjian Li View author publications You can also search for this author inPubMed Google Scholar * Xiaoliang Chen View
author publications You can also search for this author inPubMed Google Scholar * Liqun Zhu View author publications You can also search for this author inPubMed Google Scholar * Zhenghong
Lao View author publications You can also search for this author inPubMed Google Scholar * Tianhao Zhou View author publications You can also search for this author inPubMed Google Scholar *
Lijuan Zang View author publications You can also search for this author inPubMed Google Scholar * Weiyu Ge View author publications You can also search for this author inPubMed Google
Scholar * Mengyi Jiang View author publications You can also search for this author inPubMed Google Scholar * Jingxuan Xu View author publications You can also search for this author
inPubMed Google Scholar * Yuan Cao View author publications You can also search for this author inPubMed Google Scholar * Shaoqian Du View author publications You can also search for this
author inPubMed Google Scholar * Yue Yu View author publications You can also search for this author inPubMed Google Scholar * Guangjian Fan View author publications You can also search for
this author inPubMed Google Scholar * Hongxia Wang View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Guangjian Fan
or Hongxia Wang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 SUPPLEMENTARY FIGURE 2 SUPPLEMENTARY FIGURE 3 SUPPLEMENTARY FIGURE 4
SUPPLEMENTARY FIGURE 5 SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY TABLE 3 SUPPLEMENTARY TABLE 4 SUPPLEMENTARY TABLE 5 SUPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY TABLE LEGENDS
RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Li, J., Chen, X., Zhu, L. _et al._ SOX9 is a critical regulator of TSPAN8-mediated metastasis in pancreatic cancer. _Oncogene_ 40, 4884–4893 (2021).
https://doi.org/10.1038/s41388-021-01864-9 Download citation * Received: 04 November 2020 * Revised: 30 April 2021 * Accepted: 25 May 2021 * Published: 23 June 2021 * Issue Date: 29 July
2021 * DOI: https://doi.org/10.1038/s41388-021-01864-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative