- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In chronic lymphocytic leukemia (CLL), _TP53_ gene defects, due to deletion of the 17p13 locus and/or mutation(s) within the _TP53_ gene, are associated with resistance to
chemoimmunotherapy and a particularly dismal clinical outcome. On these grounds, analysis of _TP53_ aberrations has been incorporated into routine clinical diagnostics to improve patient
stratification and optimize therapeutic decisions. The predictive implications of _TP53_ aberrations have increasing significance in the era of novel targeted therapies, i.e., inhibitors of
B-cell receptor (BcR) signaling and anti-apoptotic BCL2 family members, owing to their efficacy in patients with _TP53_ defects. In this report, the _TP53_ Network of the European Research
Initiative on Chronic Lymphocytic Leukemia (ERIC) presents updated recommendations on the methodological approaches for _TP53_ mutation analysis. Moreover, it provides guidance to ensure
that the analysis is performed in a timely manner for all patients requiring treatment and that the data is interpreted and reported in a consistent, standardized, and accurate way. Since
next-generation sequencing technologies are gaining prominence within diagnostic laboratories, this report also offers advice and recommendations for the interpretation of _TP53_ mutation
data generated by this methodology. SIMILAR CONTENT BEING VIEWED BY OTHERS ERIC RECOMMENDATIONS FOR _TP53_ MUTATION ANALYSIS IN CHRONIC LYMPHOCYTIC LEUKEMIA—2024 UPDATE Article Open access
16 May 2024 TIME TO FIRST TREATMENT AND P53 DYSFUNCTION IN CHRONIC LYMPHOCYTIC LEUKAEMIA: RESULTS OF THE O-CLL1 STUDY IN EARLY STAGE PATIENTS Article Open access 28 October 2020 CLONAL
EVOLUTION AND CLINICAL IMPLICATIONS OF GENETIC ABNORMALITIES IN BLASTIC TRANSFORMATION OF CHRONIC MYELOID LEUKAEMIA Article Open access 14 May 2021 INTRODUCTION Chronic lymphocytic leukemia
(CLL) displays a very heterogeneous clinical behavior, therefore prognostic and predictive markers play an important role in disease management. To date, the key decision-making biomarkers
in CLL are _TP53_ gene defects: chromosomal aberrations of 17p13, in particular deletions spanning the _TP53_ locus, and _TP53_ gene mutations, both of which are associated with adverse
disease outcome due to resistance to chemoimmunotherapy [1,2,3,4]. Early studies utilizing fluorescent in situ hybridization (FISH), for the detection of cytogenetic aberrations, revealed
that CLL patients carrying del(17p) have a significantly shorter overall survival compared to patients harboring other recurrent cytogenetic abnormalities, i.e., del(11q), trisomy 12, or
del(13q) [5]. Inactivation of the _TP53_ locus due to del(17p) is frequently associated with mutation(s) on the second _TP53_ allele. However, _TP53_ mutations also occur in the absence of
del(17p) in about 5% of untreated patients and are associated with a poor outcome, similar to the disease course observed in del(17p) CLL patients [6, 7]. More specifically, approximately
90% of patients with del(17p) carry a _TP53_ mutation; conversely, only 60–70% of patients with _TP53_ mutation also harbor del(17p), as detected by FISH [8,9,10,11,12]. The clinical utility
of _TP53_ mutation analysis in CLL has been well documented by many studies [7,8,9, 11, 13], including findings from prospective clinical trials [6, 14, 15] clearly showing that patients
carrying _TP53_ defects are resistant to chemoimmunotherapy. In this context, the advent of novel treatment options inhibiting B-cell signaling and anti-apoptotic BCL2 that proved
efficacious in patients harboring _TP53_ gene disruption [16,17,18] has brought an urgent need for accurate assessment of the _TP53_ gene status in routine clinical practice with the aim of
identifying those patients who would not benefit from chemoimmunotherapy, and hence should be considered for targeted agents. _TP53_ gene assessment should always be performed prior to
initiation of the first and every subsequent line of treatment [19]. That said, a few situations exist where _TP53_ mutational analysis may not be required, e.g., when the use of
p53-independent drugs is not possible due to either patient fitness or limited market access, or when the presence of a _TP53_ alteration has already been documented. The recent introduction
of high-throughput next-generation sequencing (NGS) has led to the identification of _TP53_ mutations with a low variant allelic frequency (VAF)—usually below the detection limit of
conventional Sanger sequencing—that may be positively selected with the use of chemotherapy, ultimately leading to the expansion of an initially minor _TP53_ mutant subclone into a prevalent
refractory clone [20,21,22,23,24]. Taken together, the recent therapeutic and technological advances necessitate an update of the previously published ERIC recommendations for _TP53_
mutation analysis in CLL [19], including assessment of the current methodological approaches as well as recommendations for the interpretation of the findings and the accurate reporting of
results. An overview of the updated recommendations is provided in Table 1. PROCEDURE DESCRIPTION MATERIAL FOR _TP53_ MUTATION ANALYSIS For most CLL patients, peripheral blood (PB) is an
appropriate starting material for _TP53_ mutation analysis. Nevertheless, an important factor influencing the result is the cancer cell fraction (CCF), and this is particularly relevant in
cases with a low lymphocyte count (<10 × 109/L and/or <60–70% lymphocytes in PB). This is usually evidenced in patients with predominant lymphadenopathy and few circulating clonal
cells, i.e., small lymphocytic lymphoma. In such cases, material enriched with tumor cells such as bone marrow (BM) or lymph node biopsies may be an alternative option. PB or BM should be
collected in tubes containing an anticoagulant, such as EDTA or heparin, followed by mononuclear cell separation by density gradient centrifugation to enrich the lymphocyte fraction. The use
of mononuclear cells might be insufficient when the specimen analyzed contains less than 60–70% lymphocytes and could lead to a false-negative result when using Sanger sequencing
(Supplementary Fig. S1). In such instances, selection of CD19+ cells using enrichment techniques such as RosetteSep or MACS should be performed to yield a higher CCF. Alternatively,
ultra-deep NGS, which has a much greater sensitivity level, can be performed and the VAFs corrected with respect to the CCF. Regarding tissue material, fresh/frozen material is strongly
preferred. Formalin-fixed, paraffin-embedded (FFPE) tissues are recommended only when no alternative sample is available as the fixation and embedding processes may hamper the analysis,
since: (i) FFPE material often contains highly degraded DNA fragments, therefore shorter amplicons are required for sequencing; (ii) the process of tissue fixation damages DNA through
cross-linking, thus reducing the number of intact DNA molecules added into the PCR [25]; and (iii) DNA can be chemically modified, leading to artefactual sequencing results (particularly
deamination and oxidation artifacts) [26,27,28]. Therefore, any variants detected in DNA samples from FFPE material should be confirmed by independent PCR and carefully verified using the
recommended databases (described below) before interpreting and reporting them as mutations. Finally, when considering the type of nucleic acid to analyze, genomic DNA is highly recommended.
Analyzing RNA may result in truncating or splice site variants being missed due to nonsense-mediated RNA decay [29]. In addition, using whole-genome amplification for diagnostic purposes is
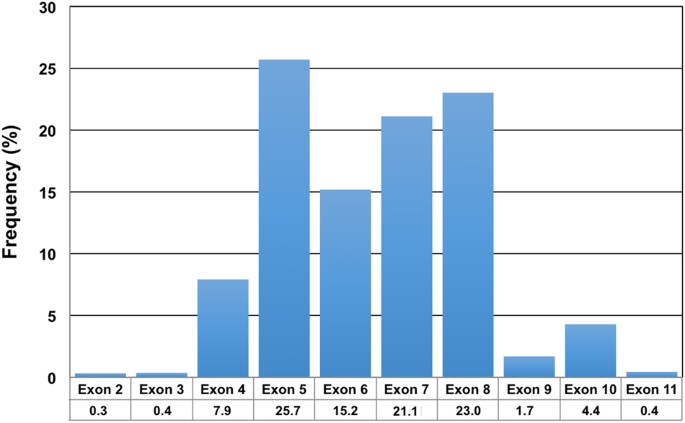
discouraged as it may introduce a bias in allelic frequencies and could lead to allelic drop-out. REGION OF INTEREST At a minimum, the sequenced region of the _TP53_ gene must include exons
4–10, which corresponds to the DNA-binding domain (codons 100–300) and the oligomerization domain (codons 323–356). Sequencing of exon 10 is recommended as the frequency of mutations in
exons 9 and 10 is similar or even higher in exon 10 as documented by the recent studies [30] (Fig. 1). Optimally, exons 2–11 should be analyzed to cover the entire coding region [30]. _TP53_
gene profiling studies by NGS, which usually involves also exons 2, 3, and 11, have shown that variants can also occur in these exons, although their frequency is low (T. Soussi,
unpublished results; Fig. 1). As each exon is surrounded by a splice donor and a splice acceptor site, sequencing of +2/−2 intronic nucleotides is required to detect variants which may
impair splicing and translate to inactive proteins. SANGER SEQUENCING Primer sequences, as well as the protocol for performing the PCR, are available on the International Agency for Research
on Cancer (IARC) _TP53_ website (http://p53.iarc.fr/ProtocolsAndTools.aspx). This PCR protocol is adaptable and can be modified based on local experience. Bidirectional sequencing analysis
is the only acceptable strategy, and the chromatograms generated by Sanger sequencing should be carefully scrutinized to ensure that somatic variants present at lower allelic frequencies are
not overlooked; adjusting software settings to detect germline homozygous and heterozygous variants is not sufficient. The ERIC _TP53_ Network provides the opportunity to analyze Sanger
sequencing data via a web-based tool called GLASS [31]. This software was purpose-built to assist with the assessment of somatic gene variations and provides a standardized variant output as
recommended by the Human Genome Variation Society (HGVS). GLASS was specifically developed to support ERIC _TP53_ Network activities and is freely accessible at
http://bat.infspire.org/genomepd/glass/ or via the ERIC website (http://www.ericll.org/guidance-toolstp53/). Finally, although the relevance of pre-screening methods, such as denaturing
high-performance liquid chromatography and high-resolution melting analysis is decreasing, they remain a viable and cost-effective option. That notwithstanding, in order to identify the
specific variant, aberrant screening results must always be confirmed by Sanger sequencing in an independent PCR. NEXT-GENERATION SEQUENCING Targeted NGS can be used for the analysis of the
_TP53_ gene as a standalone assay or as part of a gene panel investigating several genes. Numerous commercially available ready-to-use analytical kits include the _TP53_ gene, and ERIC is
conducting a multi-center collaborative effort to assess and compare various pre-designed and custom gene panel technologies. Previous studies exploring the inter-reproducibility of targeted
NGS and Sanger sequencing for _TP53_ analysis demonstrated very good correlation of the results, specifically showing that all variants detected by Sanger sequencing are also detectable by
NGS [22, 23, 32,33,34,35]. A recent study also showed an excellent correlation between the results obtained from two different NGS platforms, namely, the Ion PGM (ThermoFisher) and the MiSeq
(Illumina) [33]. In addition, NGS is capable of detecting variants below the sensitivity threshold of Sanger sequencing, even VAFs as low as <1% [20, 22, 23]. Due to the low detection
limit of NGS, multiple subclonal mutations within the _TP53_ gene (i.e., convergent mutations) may be detected in some patients [20, 35]. To ensure the maximum applicability and reliability
of NGS, several important issues need to be addressed when establishing the methodology, as erroneous results can arise for various reasons (Table 2). DNA INPUT AND QUALITY Low input and/or
degraded DNA may result in false-negative results due to a sampling effect, and may also produce false-positive results as amplified errors might constitute a significant proportion of the
final sequencing library [36]. The initial amount of DNA should always be calculated with respect to the required limit of detection (LOD), keeping in mind that a human cell (two alleles)
contains approximately 6 pg of DNA. For reliable detection, the DNA input must ensure that the sample contains a sufficient number of variant molecules and that the variants can be
distinguished from background noise. For instance, at least 10 ng corresponding to approximately 1500 cells or 3000 alleles should be used to detect variants present at 1% VAF. This is also
relevant for techniques which require the starting amount of DNA to be distributed amongst individual nano-scale PCRs, e.g., the Fluidigm Access Array, RainDance Technology, or Wafergen.
Although DNA isolated from PB and BM is usually of good quality, testing the integrity of the DNA by agarose electrophoresis or specialized automated electrophoresis devices is recommended
(and often required) for NGS. Special attention is required when considering the quality and quantity of DNA obtained from FFPE samples due to the increased risk of false-positive as well as
false-negative results. LIBRARY PREPARATION Both amplicon-based and capture-based approaches are applicable. From a practical perspective, amplicon-based library preparations require much
smaller quantities of input DNA and the workflow tends to be simpler and less time-intensive and labor-intensive compared to capture-based methodologies. On the other hand, hybridization
capture-based approaches demonstrate better uniformity of coverage and generate fewer false-negative as well as false-positive calls as compared to amplicon-based techniques. When designing
in-house primers for amplicon-based libraries, it is important to check the primer positions against potential single-nucleotide polymorphisms (SNP) and ensure that the primers can
efficiently read across splice junctions. In order to establish an NGS assay with high detection sensitivity, proofreading polymerases with low error-rates are recommended. Incorporating
unique molecular identifiers into the library preparation helps to distinguish errors introduced artificially during the process from true low-frequency variants and also allows for more
accurate quantification (especially with PCR-based protocols) [37, 38]. Additional benchmarking studies are required to establish standard analytical methods that must then be checked for
accuracy and reproducibility. SEQUENCING AND COVERAGE The required coverage should be set to ensure that the call is statistically above the background noise. Generally, the minimal coverage
should not be less than 100 at any position within the regions of interest and the number of variant reads for reliable variant calling should be at least 10. The frequently reported mean
or median coverage of a diagnostic panel is non-informative as uncovered regions cannot be deduced from this average value and therefore a ≥99% minimum coverage percentage is a vital
requirement. Of note, the number of reads does not necessarily reflect the actual number of unique template gDNA molecules, as many reads will be duplicates generated during PCR
amplification. When employing longer reads, a confident overlap (>60–70%) between the paired reads is recommended in order to avoid the introduction of false-positive results. Calling
variants found in unbalanced regions with forward-reverse ratios of less than 10% (i.e., strand bias) should be avoided. DATA ANALYSIS Multiple commercial, as well as free, software tools
are available to analyze NGS data and, as the bioinformatics field is continuously evolving, no single tool is currently preferentially recommended. That said, it is of utmost importance to
use a pipeline that has been optimized, and validated, for the detection of low abundance variants that must be distinguished from background error noise. Another issue concerns the accurate
identification of insertions and deletions (indels), which may be missed during the alignment process, especially in the case of complex indels. Numerous indel-calling tools have been
developed that often vary in the manner by which they detect indel breakpoints. Performance evaluations of indel-calling software have revealed limitations in detection; consequently, manual
inspection of the data is always recommended and is particularly required for indel variants and variants close to the detection limit. LIMIT OF DETECTION LOD refers to the lowest VAF that
is reproducibly detectable by the particular method under specific well-defined conditions. The LOD is a function of both the initial DNA input and the coverage achieved. The NGS assay
should be established, and validated, to at least reliably identify variants detectable by Sanger sequencing and avoid false-positive calls with VAF above the Sanger sequencing detection
limit (e.g., minimum LOD is 10% VAF). LOD should be set by taking into account non-uniformity of coverage across the analyzed sequence and an inconsistent error distribution. The occurrence
of sequencing errors varies depending on the nucleotide position and composition and is also platform-dependent, with C:G>T:A being the most frequent using Illumina platforms [39]. The
error rate is also influenced by the specific sequence context (e.g., homopolymers are more prone to erroneous variant calling). The issue of detection limit and how it can influence the
interpretation of findings is discussed in the following section. CLINICAL REPORTING AND INTERPRETATION OF THE RESULTS VARIANT DESCRIPTION Detected variants should be described using the
nomenclature devised by the HGVS nomenclature (http://varnomen.hgvs.org/) [40]. Several software programs are available to ensure adherence to standardized nomenclature (e.g., Mutalyzer;
https://www.mutalyzer.nl/). Variants should be described at both cDNA and protein level, and the reference sequence number and version including the transcript and protein variant should be
stated (see Supplementary Material). To standardize the output, the preferred coding DNA reference sequence is the stable Locus Reference Genomic sequence (LRG;
http://ftp.ebi.ac.uk/pub/databases/lrgex/LRG_321.xml) [30]. Transcript and protein variants 1 should be used (LRG_321t1, LRG_321p1). Special attention is warranted when annotating variants
detected by NGS, especially since many bioinformatics pipelines do not fulfill the requirements for correct variant description according to the HGVS nomenclature. More specifically: (i)
insertions and deletions are often not handled accurately; (ii) duplications are often misinterpreted as insertions; (iii) varying reference sequences for _TP53_ within the same output are
used; and (iv) the 3′ rule is not always implemented correctly. This is of particular importance for _TP53_ and other genes that are oriented in the reverse direction on the chromosome. In
such situations, the alignment and variant calling steps may introduce errors if aligning to the 3′ end with respect to the chromosome position rather than the coding sequence orientation.
INTERPRETATION DATABASES The detected variant should be checked using locus-specific databases, i.e., either the IARC _TP53_ database (http://p53.iarc.fr/TP53GeneVariations.aspx) [41] or the
_TP53_ website (UMD database; http://p53.fr/) [42]. These databases compile data from peer-reviewed literature as well as general databases, and provide information about: (i) the
functional impact of all possible single-nucleotide exchanges within the coding region; (ii) the variant frequencies noted in both the somatic and the germline context; and (iii) additional
relevant information, including links to other resources. The _TP53_ website also provides a web-service tool called Seshat that is capable of managing files generated from NGS both in the
vcf and bam formats. Seshat helps the user to: (i) check the variant nomenclature for consistency and generate a full description of each variant formatted according to HGVS; (ii) assess the
pathogenicity of each variant according to general prediction algorithms and algorithms developed specifically for analyzing the _TP53_ gene; and (iii) obtain functional and structural data
for each _TP53_ variant. Finally, variants can also be checked using the COSMIC (http://cancer.sanger.ac.uk/cosmic) or ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) databases; however,
these databases are only recommended as a complementary analysis to the locus-specific databases. POLYMORPHISMS AND NEUTRAL VARIANTS In general, it is not recommended to include common
polymorphisms and benign variants in the report to physicians. If, however, the local practice requires that these variants are detailed in the clinical report, it should be clearly
indicated that the detected variant is not clinically relevant. According to the IARC database, there are six validated exonic polymorphisms within the _TP53_ gene; two are synonymous
(c.108G>A: p.Pro36= and c.639A>G: p.Arg213=) and four are nonsynonymous (c.91G>A: p.Val31Ile; c.139C>T: p.Pro47Ser; c.215C>G: p.Pro72Arg, and c.1096T>G: p.Ser366Ala). The
most frequent polymorphism is c.215C>G: p.Pro72Arg, where the ancestral allele C coding for proline is less frequent in the general population than the allele G [43] with
latitude-dependent variations. Although the two alleles were reported to have different capabilities in inducing apoptosis and G1 arrest [44], studies analyzing the clinical impact of
p.Pro72Arg and its associations with _TP53_ mutations in CLL reported inconclusive results [45,46,47,48]. Reporting of the p.Pro72Arg status is therefore not recommended due to a lack of
convincing evidence with regard to prognostic or clinical relevance. Using dbSNP for filtering out polymorphisms and neutral variants is strongly discouraged as many variants listed in dbSNP
exhibit loss of function and are frequently observed in cancer patients despite not being reported as pathogenic in ClinVar [49]. More specifically, of the 100 most frequent deleterious
somatic variants described in the IARC database, 65 are present in dbSNP147 and only 34 are described as being pathogenic [41]. Using the data set collected within the context of the Genome
Aggregation Database (gnomAD) is more accurate; however, it should be noted that several pathological variants are also listed in this database (http://gnomad.broadinstitute.org/, originally
Exome Aggregation Consortium [43]). VARIANTS WITH PRESERVED ACTIVITY If a rare variant or a variant with preserved functionality is detected, it is recommended to repeat the entire
analysis, starting from the PCR step, so as to exclude analytical errors. If the variant is verified and the VAF is approximately 50%, suggesting a germline origin, it is advisable to verify
the germline or somatic nature of the variant by testing patient-matched germline DNA, obtained from CD3+ cells, saliva, a buccal swab or a skin biopsy (it is advised to rule out the
contamination with CLL cells by flow cytometry or by testing the patient-specific IGHV rearrangement). Variants that have preserved transactivation capabilities are often found as germline
and the carriers do not show any personal or family cancer-history associated with Li-Fraumeni or another cancer-predisposing syndrome. Specific examples of variants that should be
considered with caution and are often inaccurately reported are c.704A>G: p.Asn235Ser or c.847C>T: p.Arg283Cys. If the somatic origin of such a variant is confirmed, the variant should
be reported to the clinician clearly stating that a variant of unknown significance was found. In the case that the variant is of germline origin, reporting should follow the
recommendations of The American College of Medical Genetics and Genomics [50, 51] (recommendations of The European Society of Human Genetics are currently under preparation). INTRONIC
VARIANTS Variants affecting splice sites (+2/−2 intronic nucleotides) are considered pathogenic as they lead to aberrant mRNA splicing. Pathogenicity of intronic variants outside the donor
and acceptor sequence is largely unexplored, and therefore they should not be reported unless their functional impact is proven at the RNA or protein level by documenting the presence of
aberrantly spliced transcripts or shortened protein products. As these methods are not usually accessible in diagnostic labs, reporting of intronic variants with the exception of splice
sites is not recommended within clinical routine. SYNONYMOUS VARIANTS If a synonymous variant is detected, it is important to check its predicted effect on splicing [52] via the IARC
database or the _TP53_ website. For instance, synonymous variants in codon 125 (c.375G>A and c.375G>T) have been found in various cancers and Li-Fraumeni families and shown to affect
the splicing of exon 4 [53], therefore they are classified as pathogenic. INDEL VARIANTS Insertions and deletions leading to the formation of a premature stop codon (frameshift variants) as
well as in-frame indels within the DNA-binding domain are considered as likely pathogenic. CLINICAL REPORTING OF SUBCLONAL VARIANTS WITH LOW VARIANT ALLELE FREQUENCY DETECTED BY NGS The
definition of the term “subclonal” is generally used to describe variants that are not present in the entire tumor population, as opposed to “clonal” [21]. Terms such as “minor subclone”,
“low-burden”, or “low-level” variants refer to variants with allelic burdens below the detection limit of Sanger sequencing, i.e., <10% VAF. Of note, caution is necessary when
interpreting VAFs as its calculation does not take into consideration the CCF and the presence of genomic copy number aberrations. Therefore, it is important to bear in mind that a 5% VAF
could be clonal if the CCF is only 10% and no del(17p) or copy-neutral loss of heterozygosity is present. Several publications have suggested that _TP53_ mutations within minor clones are
clinically relevant, which is particularly important considering that administration of therapeutic regimens based on DNA-damaging agents represents a risk for the selection of these
low-level _TP53_-mutated subclones [20,21,22,23, 33, 54]. However, the extent of the risk posed by minor subclones harboring _TP53_ mutations has not been conclusively defined, and the
current evidence on the poor outcome of _TP53_-mutated patients treated with chemoimmunotherapy in clinical trials is based on data obtained using Sanger sequencing only. Therefore,
currently, the presence of minor subclonal mutations should not impact clinical decision-making. Based on current knowledge, the recommended threshold for reporting of mutations detected by
NGS should reflect the Sanger-like threshold of approximately ~10% VAF. That said, bearing in mind that the 10% threshold is arbitrary, variants with 5–10% VAF can also be reported; however,
always mentioning in the report that the clinical significance of _TP53_ mutations with VAF 5–10% is currently unknown, since we are lacking data from prospective clinical studies
addressing this issue. Importantly, NGS technology should be validated to a LOD above which there are no false positives (minimum 10% VAF). Confirmation of mutations detected at the level
near the validated LOD is desirable either by Sanger sequencing or, in the case of minor clone variants, by digital PCR, independent NGS run or allele-specific PCR. REPORT FORM In addition
to the obligatory standard medical report content (e.g., patient and lab identifiers, date of sampling, type of material), the report should always contain the following information: (i) the
type of analysis and description of the method: methodology used, exons analyzed, LOD, and coverage in the case of NGS (median and ≥99% minimum); (ii) results and interpretation:
description of the identified variant(s) according to the HGVS nomenclature, reference sequence used, type of variant (missense/truncating etc.), effect according to the _TP53_
locus-specific database, frequency, and any known association with cancer; (iii) conclusion: clinical consequence of the variant and summary of the finding in the context of the current
knowledge; and (iv) other optional data: VAF of the detected variant if available (estimations from Sanger sequence traces can also be informative), comparison with a previously tested
sample from the same patient and, if evidenced, description of clonal evolution. All labs issuing clinical reports of their results must have accreditation according to their national
authorities. ERIC is also regularly conducting _TP53_ mutational Analysis Certification to confirm the reliability and reproducibility of the results provided by participating labs. Examples
of report forms for both Sanger sequencing and NGS are provided in the Supplementary Material and a template report form can be found on the ERIC website (http://www.ericll.org/).
PUBLISHING AND SCIENTIFIC REPORTING IN THE DATABASES It is important to distinguish between clinical reporting and reporting variants for research purposes in scientific journals. Data from
publications are transferred to databases, and these databases then serve as the source of information for general use [42, 49]. For this reason, in order to prevent incorrect entries, it is
essential to follow specific rules in addition to all above-mentioned basic procedures: (i) using consistent sample and patient identifiers if the data are repeatedly published, as
inconsistent identification leads to redundancy in mutation databases; (ii) including the genomic coordinate and reference genome in the variant description to avoid ambiguities; (iii)
listing all variants that are found in the patient including synonymous and other benign variants [55]. It is recommended to include the complete list of variants in the Supplementary
Material, with appropriate description of their clinical significance. Note that if more than one variant in a patient is found, all variants should be listed. Centers following ERIC
recommendations are kindly asked to mention ERIC in the “Material and methods” section of their studies and refer to this manuscript. CONCLUDING REMARKS In CLL, inactivation of the _TP53_
gene by deletion and/or mutation is strongly associated with adverse prognosis and refractoriness to chemoimmunotherapy. Detection of del(17p) and _TP53_ gene mutations has become an
integral part in routine diagnostics and should always be performed before deciding about treatment. Analysis of _TP53_ exons 4–10 is a minimal requirement; however, ideally, the entire
coding sequence, i.e., exons 2–11, should be analyzed, and this can be performed by either bidirectional Sanger sequencing or NGS. NGS also allows the parallel analysis of multiple genes and
is capable of identifying variants undetectable by Sanger sequencing. That notwithstanding, NGS currently faces certain technical limitations and may lead to problems with data
interpretation. The clinical importance of mutations within minor clones remains an unresolved issue and there is currently not enough evidence for making therapeutic decisions based on the
presence of mutations undetectable by Sanger sequencing. To assist the community with the implementation of _TP53_ mutational analysis in a harmonized manner, ERIC created the _TP53_ Network
with the following objectives: regular certification of laboratories for _TP53_ mutation status assessment (both for Sanger and NGS), the organization of educational events, and regular
updating of recommendations for _TP53_ analysis. The Network also provides tools facilitating laboratories to achieve reliable and comparable results that are accessible via the ERIC web
page (http://www.ericll.org/). REFERENCES * Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients
with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74. Article CAS Google Scholar * Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi
M, Döhner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54. Article CAS Google Scholar * Fischer K,
Cramer P, Busch R, Böttcher S, Bahlo J, Schubert J, et al. Bendamustine in combination with rituximab for previously untreated patients with chronic lymphocytic leukemia: a multicenter phase
II trial of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. 2012;30:3209–16. Article CAS Google Scholar * Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero
G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the
National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. Article CAS Google Scholar * Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al.
Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. Article CAS Google Scholar * Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D,
et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–9. Article Google Scholar * Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, et al.
The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res.
2009;15:995–1004. Article CAS Google Scholar * Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic
lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–9. Article CAS Google Scholar * Malcikova J, Smardova J, Rocnova L,
Tichy B, Kuglik P, Vranova V, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood.
2009;114:5307–14. Article CAS Google Scholar * Zainuddin N, Murray F, Kanduri M, Gunnarsson R, Smedby KE, Enblad G, et al. TP53 mutations are infrequent in newly diagnosed chronic
lymphocytic leukemia. Leuk Res. 2011;35:272–4. Article CAS Google Scholar * Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, et al. The detection of TP53 mutations in chronic
lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–24. Article CAS Google Scholar *
Zenz T, Häbe S, Denzel T, Mohr J, Winkler D, Bühler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the
contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. Article CAS Google Scholar * Stengel A, Kern W,
Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia.
2017;31:705–11. Article CAS Google Scholar * Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and
survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. Article Google Scholar * Stilgenbauer S, Schnaiter A, Paschka P, Zenz
T, Rossi M, Dohner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54. Article CAS Google Scholar *
Stilgenbauer S, Eichhorst B, Schetelig J, Coutre S, Seymour JF, Munir T, et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre,
open-label, phase 2 study. Lancet Oncol. 2016;17:768–78. Article CAS Google Scholar * O’Brien S, Jones JA, Coutre SE, Mato AR, Hillmen P, Tam C, et al. Ibrutinib for patients with
relapsed or refractory chronic lymphocytic leukaemia with 17p deletion (RESONATE-17): a phase 2, open-label, multicentre study. Lancet Oncol. 2016;17:1409–18. Article Google Scholar *
Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic
leukemia. Blood. 2014;123:3390–7. Article CAS Google Scholar * Pospisilova S, Gonzalez D, Malcikova J, Trbusek M, Rossi D, Kater AP, et al. ERIC recommendations on TP53 mutation analysis
in chronic lymphocytic leukemia. Leukemia. 2012;26:1458–61. Article CAS Google Scholar * Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N, et al. Detailed analysis of
therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29:877–85. Article CAS Google Scholar * Landau DA, Carter SL, Stojanov P, McKenna A,
Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. Article CAS Google Scholar * Nadeu F, Delgado J, Royo
C, Baumann T, Stankovic T, Pinyol M, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1 and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–30.
Article CAS Google Scholar * Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia.
Blood. 2014;123:2139–47. Article CAS Google Scholar * Zenz T, Habe S, Denzel T, Winkler D, Dohner H, Stilgenbauer S. How little is too much? p53 inactivation: from laboratory cutoff to
biological basis of chemotherapy resistance. Leukemia. 2008;22:2257–8. Article CAS Google Scholar * Lin MT, Mosier SL, Thiess M, Beierl KF, Debeljak M, Tseng LH, et al. Clinical
validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141:856–66. Article CAS Google Scholar * Oh E, Choi YL, Kwon MJ, Kim RN, Kim
YJ, Song JY, et al. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples. PLoS ONE. 2015;10:e0144162. Article Google
Scholar * Williams C, Pontén F, Moberg C, Söderkvist P, Uhlén M, Pontén J, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol.
1999;155:1467–71. Article CAS Google Scholar * Edlund K, Larsson O, Ameur A, Bunikis I, Gyllensten U, Leroy B, et al. Data-driven unbiased curation of the TP53 tumor suppressor gene
mutation database and validation by ultradeep sequencing of human tumors. Proc Natl Acad Sci USA. 2012;109:9551–6. Article CAS Google Scholar * Lykke-Andersen S, Jensen TH.
Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–77. Article CAS Google Scholar * Leroy B, Ballinger ML, Baran-Marszak
F, Bond GL, Braithwaite A, Concin N, et al. Recommended guidelines for validation, quality control, and reporting of TP53 variants in clinical practice. Cancer Res. 2017;77:1250–60. Article
CAS Google Scholar * Pal K, Bystry V, Reigl T, Demko M, Krejci A, Touloumenidou T, et al. GLASS: assisted and standardized assessment of gene variations from Sanger sequence trace data.
Bioinformatics. 2017;33:3802–4. Article CAS Google Scholar * Kantorova B, Malcikova J, Smardova J, Pavlova S, Trbusek M, Tom N, et al. TP53 mutation analysis in chronic lymphocytic
leukemia: comparison of different detection methods. Tumour Biol. 2015;36:3371–80. Article CAS Google Scholar * Lazarian G, Tausch E, Eclache V, Sebaa A, Bianchi V, Letestu R, et al. TP53
mutations are early events in chronic lymphocytic leukemia disease progression and precede evolution to complex karyotypes. Int J Cancer. 2016;139:1759–63. Article CAS Google Scholar *
Sutton LA, Ljungström V, Mansouri L, Young E, Cortese D, Navrkalova V, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach
will facilitate implementation in a clinical setting. Haematologica. 2015;100:370–6. Article CAS Google Scholar * Jethwa A, Hüllein J, Stolz T, Blume C, Sellner L, Jauch A, et al.
Targeted resequencing for analysis of clonal composition of recurrent gene mutations in chronic lymphocytic leukaemia. Br J Haematol. 2013;163:496–500. Article CAS Google Scholar * Akbari
M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7:36–9.
Article CAS Google Scholar * Hiatt JB, Pritchard CC, Salipante SJ, O’Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency
variation. Genome Res. 2013;23:843–54. Article CAS Google Scholar * Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively
parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–5. Article Google Scholar * Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR. Cytosine deamination is a major cause of baseline
noise in next-generation sequencing. Mol Diagn Ther. 2014;18:587–93. Article CAS Google Scholar * den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al.
HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–9. Article Google Scholar * Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil
J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37:865–76. Article CAS Google Scholar * Leroy B, Anderson M, Soussi
T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672–88. Article CAS Google Scholar * Lek M, Karczewski KJ, Minikel EV,
Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. Article CAS Google Scholar * Dumont P, Leu JI, Della Pietra
AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. Article CAS Google Scholar * Kochethu G, Delgado
J, Pepper C, Starczynski J, Hooper L, Krishnan S, et al. Two germ line polymorphisms of the tumour suppressor gene p53 may influence the biology of chronic lymphocytic leukaemia. Leuk Res.
2006;30:1113–8. Article CAS Google Scholar * Majid A, Richards T, Dusanjh P, Kennedy DB, Miall F, Gesk S, et al. TP53 codon 72 polymorphism in patients with chronic lymphocytic leukaemia:
identification of a subgroup with mutated IGHV genes and poor clinical outcome. Br J Haematol. 2011;153:533–5. Article Google Scholar * Dong HJ, Fang C, Wang L, Fan L, Xu J, Wu JZ, et al.
TP53 Pro72 allele potentially increases the poor prognostic significance of TP53 mutation in chronic lymphocytic leukemia. Med Oncol. 2014;31:908. Article Google Scholar * Sturm I,
Bosanquet AG, Hummel M, Dörken B, Daniel PT. In B-CLL, the codon 72 polymorphic variants of p53 are not related to drug resistance and disease prognosis. BMC Cancer. 2005;5:105. Article
Google Scholar * Soussi T, Leroy B, Taschner PE. Recommendations for analyzing and reporting TP53 gene variants in the high-throughput sequencing era. Hum Mutat. 2014;35:766–78. Article
CAS Google Scholar * Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016
update (ACMG SFv2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55. Article Google Scholar * Green RC, Berg JS, Grody WW, Kalia
SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. Article CAS Google Scholar *
Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–35. Article CAS Google Scholar * Varley
JM, Attwooll C, White G, McGown G, Thorncroft M, Kelsey AM, et al. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene. 2001;20:2647–54.
Article CAS Google Scholar * Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature.
2015;526:525–30. Article CAS Google Scholar * Soussi T, Taschner PE, Samuels Y. Synonymous somatic variants in human cancer are not infamous: a plea for full disclosure in databases and
publications. Hum Mutat. 2017;38:339–42. Article CAS Google Scholar Download references FUNDING Supported by the IMI 2 HARMONY JU under GA No 116026, this JU receives support from the
EU’s H2020 R&I program and EFPIA. Further supported by the EU Horizon 2020 projects MEDGENET 692298, AEGLE 644906, projects CEITEC 2020 (LQ1601), NCMG research infrastructure (LM2015091
funded by MEYS CR), project FNBr 65269705, FM MU ROZV/24/LF/2016, DFG (SFB1074, projects B1 and B2, and EU (FIRE CLL)), and the Swedish Cancer Society and the Swedish Research Council.
Publication reflects only the authors’ views and the Commission is not responsible for any use that may be made of the information it contains. AUTHOR INFORMATION Author notes * J.
Malcikova, E. Tausch, and D. Rossi contributed equally to this work * P. Ghia and S. Pospisilova contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Internal Medicine
— Hematology and Oncology, University Hospital Brno and Medical Faculty, Masaryk University, Brno, Czech Republic J. Malcikova & S. Pospisilova * Central European Institute of
Technology, Masaryk University, Brno, Czech Republic J. Malcikova & S. Pospisilova * Department of Internal Medicine III, Ulm University, Ulm, Germany E. Tausch & S. Stilgenbauer *
Hematology, Oncology Institute of Southern Switzerland, Institute of Oncology Research, Bellinzona, Switzerland D. Rossi * Department of Immunology, Genetics and Pathology, Science for Life
Laboratory, Uppsala University, Uppsala, Sweden L. A. Sutton & R. Rosenquist * Department of Molecular Medicine and Surgery, Karolinska Institutet, Stockholm, Sweden L. A. Sutton &
R. Rosenquist * Université Pierre et Marie Curie, Paris, France T. Soussi * INSERM, U1138, Centre de Recherche des Cordeliers, Paris, France T. Soussi * Department of Oncology-Pathology,
Karolinska Institutet, Cancer Center Karolinska, Stockholm, Sweden T. Soussi * Division of Hematology, University Hospital Zürich, University of Zürich, Zürich, Switzerland T. Zenz *
Department of Hematology, Academic Medical Center, Amsterdam, The Netherlands A. P. Kater * Department of Hematology, Rigshospitalet, Copenhagen, Denmark C. U. Niemann * Centre for Cancer
Research and Cell Biology, Queen’s University Belfast, Belfast, UK D. Gonzalez * Department of Hematology, Hôpital Pitié-Salpêtière, AP-HP, Sorbonne Universités-UPMC University, Paris,
France F. Davi * Centro de Investigación del Cancer and Centro de Investigación Biomédica en Red de Cáncer (CIBERONC), University of Salamanca, Salamanca, Spain M. Gonzalez Diaz * Department
of Haematology, Hospital de la Santa Creu I Sant Pau, Autonomous University of Barcelona, Barcelona, Spain C. Moreno * Division of Haematology, Department of Translational Medicine,
University of Eastern Piedmont, Novara, Italy G. Gaidano * Institute of Applied Biosciences, CERTH, Thessaloniki, Greece K. Stamatopoulos * Division of Experimental Oncology, IRCCS San
Raffaele Scientific Institute, Università Vita-Salute San Raffaele, Milan, Italy P. Ghia Authors * J. Malcikova View author publications You can also search for this author inPubMed Google
Scholar * E. Tausch View author publications You can also search for this author inPubMed Google Scholar * D. Rossi View author publications You can also search for this author inPubMed
Google Scholar * L. A. Sutton View author publications You can also search for this author inPubMed Google Scholar * T. Soussi View author publications You can also search for this author
inPubMed Google Scholar * T. Zenz View author publications You can also search for this author inPubMed Google Scholar * A. P. Kater View author publications You can also search for this
author inPubMed Google Scholar * C. U. Niemann View author publications You can also search for this author inPubMed Google Scholar * D. Gonzalez View author publications You can also search
for this author inPubMed Google Scholar * F. Davi View author publications You can also search for this author inPubMed Google Scholar * M. Gonzalez Diaz View author publications You can
also search for this author inPubMed Google Scholar * C. Moreno View author publications You can also search for this author inPubMed Google Scholar * G. Gaidano View author publications You
can also search for this author inPubMed Google Scholar * K. Stamatopoulos View author publications You can also search for this author inPubMed Google Scholar * R. Rosenquist View author
publications You can also search for this author inPubMed Google Scholar * S. Stilgenbauer View author publications You can also search for this author inPubMed Google Scholar * P. Ghia View
author publications You can also search for this author inPubMed Google Scholar * S. Pospisilova View author publications You can also search for this author inPubMed Google Scholar
CONSORTIA ON BEHALF OF THE EUROPEAN RESEARCH INITIATIVE ON CHRONIC LYMPHOCYTIC LEUKEMIA (ERIC) — TP53 NETWORK CORRESPONDING AUTHORS Correspondence to P. Ghia or S. Pospisilova. ETHICS
DECLARATIONS CONFLICT OF INTEREST JM and SP: consultancy fees and travel grants from Gilead and Abbvie. DR: research funding from Abbvie and Gilead, consultancy fees from Abbvie, Janssen,
Gilead. LAS: honoraria for consultancy from Gilead and Janssen. TZ: honoraria from Janssen, Gilead, Abbvie, Vaniam Group, Roche. APK: research funding from Janssen, Gilead, Abbvie, Celgene,
Roche. CN: research funding from Novo Nordisk Foundation, Danish Cancer Foundation and Abbvie and consultancy fees and/or travel grants from Roche, Janssen, Novartis, Gilead, and Abbvie. FD:
consultant fees from Gilead. CM: consultant fees from Janssen, Gilead, Pharmacyclics and research funding from Roche and Gilead. GG: consultancy fees from Janssen, Gilead, Roche, Morphosys,
and Abbvie. KS: research support from Janssen Pharmaceuticals, Gilead Sciences, Novartis SA, and Abbvie. RR: consultancy fees from Gilead and Roche. SS: honoraria for consultancy, honoraria
and research grants from AbbVie, Celgene, Genentech, Gilead, GSK, Hoffmann La-Roche, Janssen, Novartis, Pharmacyclics. PG: honoraria for consultancy and research grants from AbbVie,
Janssen, Gilead, Roche. The remaining authors declare that they have no conflict of interest. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY FIGURE 1 – ILLUSTRATIVE EXAMPLE OF A CLL PATIENT
WITH A LOW LYMPHOCYTE COUNT IN THE PERIPHERAL BLOOD. SUPPLEMENTARY FILE 1 – EXAMPLE OF A REPORT FORM: NEGATIVE RESULT, SANGER SEQUENCING SUPPLEMENTARY FILE 2 – EXAMPLE OF A REPORT FORM:
NEGATIVE RESULT, NGS SUPPLEMENTARY FILE 3 – EXAMPLE OF A REPORT FORM: HOT SPOT MUTATION, NGS SUPPLEMENTARY FILE 4 – EXAMPLE OF A REPORT FORM: 2 MUTATIONS (SPLICE SITE AND DELETION), SANGER
SEQUENCING SUPPLEMENTARY FILE 5 – EXAMPLE OF A REPORT FORM: NONSENSE MUTATION WITH LOW VAF, NGS SUPPLEMENTARY FILE 6 – EXAMPLE OF A REPORT FORM, VARIANT OF UNKNOWN SIGNIFICANCE, SANGER
SEQUENCING RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, which permits any
non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a
link to the Creative Commons license, and indicate if changes were made. If you remix, transform, or build upon this article or a part thereof, you must distribute your contributions under
the same license as the original. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line
to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/. Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Malcikova, J., Tausch, E., Rossi, D. _et al._ ERIC recommendations for _TP53_ mutation analysis in chronic lymphocytic leukemia—update on methodological approaches
and results interpretation. _Leukemia_ 32, 1070–1080 (2018). https://doi.org/10.1038/s41375-017-0007-7 Download citation * Received: 01 September 2017 * Revised: 05 December 2017 * Accepted:
08 December 2017 * Published: 02 February 2018 * Issue Date: May 2018 * DOI: https://doi.org/10.1038/s41375-017-0007-7 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative