- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT DNA double-strand breaks (DSBs) can be repaired either via homologous recombination (HR) or nonhomologous end-joining (NHEJ). Both pathways are operative in eukaryotes, but bacteria
had been thought to rely on HR alone. Here we provide direct evidence that mycobacteria have a robust NHEJ pathway that requires Ku and a specialized polyfunctional ATP-dependent DNA ligase
(LigD). NHEJ of blunt-end and complementary 5′-overhang DSBs is highly mutagenic (∼50% error rate). Analysis of the recombination junctions ensuing from individual NHEJ events highlighted
the participation of several DNA end-remodeling activities, including template-dependent fill-in of 5′ overhangs, nontemplated addition of single nucleotides at blunt ends, and nucleolytic
resection. LigD itself has the template-dependent and template-independent polymerase functions _in vitro_ that compose the molecular signatures of NHEJ _in vivo_. Another ATP-dependent DNA
ligase (LigC) provides a backup mechanism for LigD-independent error-prone repair of blunt-end DSBs. We speculate that NHEJ allows mycobacteria to evade genotoxic host defense. Access
through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal
Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
SIMILAR CONTENT BEING VIEWED BY OTHERS POLΛ PROMOTES MICROHOMOLOGY-MEDIATED END-JOINING Article 19 December 2022 THE IMPORTANCE OF DNAPKCS FOR BLUNT DNA END JOINING IS MAGNIFIED WHEN XLF IS
WEAKENED Article Open access 27 June 2022 STRUCTURAL BASIS FOR POLΘ-HELICASE DNA BINDING AND MICROHOMOLOGY-MEDIATED END-JOINING Article Open access 19 April 2025 REFERENCES * Buchmeier, N.A.
et al. DNA repair is more important than catalase for _Salmonella_ virulence in mice. _J. Clin. Invest._ 95, 1047–1053 (1995). Article CAS Google Scholar * Buchmeier, N.A., Lipps, C.J.,
So, M.Y. & Heffron, F. Recombination-deficient mutants of _Salmonella typhimurium_ are avirulent and sensitive to the oxidative burst of macrophages. _Mol. Microbiol._ 7, 933–936 (1993).
Article CAS Google Scholar * O'Rourke, E.J. et al. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in _Helicobacter pylori_
colonization. _Proc. Natl. Acad. Sci. USA_ 100, 2789–2794 (2003). Article CAS Google Scholar * Suvarnapunya, A.E., Lagasse, H.A. & Stein, M.A. The role of DNA base excision repair in
the pathogenesis of _Salmonella enterica_ serovar _Typhimurium_. _Mol. Microbiol._ 48, 549–559 (2003). Article CAS Google Scholar * Sander, P. et al. _Mycobacterium bovis_ BCG _recA_
deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. _Infect. Immun._ 69, 3562–3568 (2001). Article CAS Google Scholar
* Darwin, K.H., Ehrt, S., Gutierrez-Ramos, J.C., Weich, N. & Nathan, C.F. The proteasome of _Mycobacterium tuberculosis_ is required for resistance to nitric oxide. _Science_ 302,
1963–1966 (2003). Article CAS Google Scholar * Boshoff, H.I., Reed, M.B., Barry, C.E. & Mizrahi, V. DnaE2 polymerase contributes to _in vivo_ survival and the emergence of drug
resistance in _Mycobacterium tuberculosis_. _Cell_ 113, 183–193 (2003). Article CAS Google Scholar * Cromie, G.A., Connelly, J.C. & Leach, D.R. Recombination at double-strand breaks
and DNA ends: conserved mechanisms from phage to humans. _Mol. Cell_ 8, 1163–1174 (2001). Article CAS Google Scholar * Lieber, M.R., Ma, Y., Pannicke, U. & Schwarz, K. Mechanism and
regulation of human non-homologous DNA end-joining. _Nat. Rev. Mol. Cell Biol._ 4, 712–720 (2003). Article CAS Google Scholar * Downs, J.A. & Jackson, S.P. A means to a DNA end: the
many roles of Ku. _Nat. Rev. Mol. Cell Biol._ 5, 367–378 (2004). Article CAS Google Scholar * Riballo, E. et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia
patient. _Curr. Biol._ 9, 699–702 (1999). Article CAS Google Scholar * Barnes, D.E., Tomkinson, A.E., Lehmann, A.R., Webster, A.D. & Lindahl, T. Mutations in the DNA ligase I gene of
an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. _Cell_ 69, 495–503 (1992). Article CAS Google Scholar * Schar, P., Herrmann, G., Daly, G. &
Lindahl, T. A newly identified DNA ligase of _Saccharomyces cerevisiae_ involved in RAD52-independent repair of DNA double-strand breaks. _Genes Dev._ 11, 1912–1924 (1997). Article CAS
Google Scholar * Teo, S.H. & Jackson, S.P. Identification of _Saccharomyces cerevisiae_ DNA ligase IV: involvement in DNA double-strand break repair. _EMBO J._ 16, 4788–4795 (1997).
Article CAS Google Scholar * Wilson, T.E., Grawunder, U. & Lieber, M.R. Yeast DNA ligase IV mediates non-homologous DNA end joining. _Nature_ 388, 495–498 (1997). Article CAS Google
Scholar * Frank, K.M. et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. _Nature_ 396, 173–177 (1998). Article CAS Google Scholar *
Grawunder, U., Zimmer, D., Fugmann, S., Schwarz, K. & Lieber, M.R. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes.
_Mol. Cell_ 2, 477–484 (1998). Article CAS Google Scholar * Konrad, E.B., Modrich, P. & Lehman, I.R. Genetic and enzymatic characterization of a conditional lethal mutant of
_Escherichia coli_ K12 with a temperature-sensitive DNA ligase. _J. Mol. Biol._ 77, 519–529 (1973). Article CAS Google Scholar * Magnet, S. & Blanchard, J.S. Mechanistic and kinetic
study of the ATP-dependent DNA ligase of _Neisseria meningitidis_. _Biochemistry_ 43, 710–717 (2004). Article CAS Google Scholar * Wilkinson, A., Day, J. & Bowater, R. Bacterial DNA
ligases. _Mol. Microbiol._ 40, 1241–1248 (2001). Article CAS Google Scholar * Cheng, C. & Shuman, S. Characterization of an ATP-dependent DNA ligase encoded by _Haemophilus
influenzae_. _Nucleic Acids Res._ 25, 1369–1374 (1997). Article CAS Google Scholar * Gong, C., Martins, A., Bongiorno, P., Glickman, M. & Shuman, S. Biochemical and genetic analysis
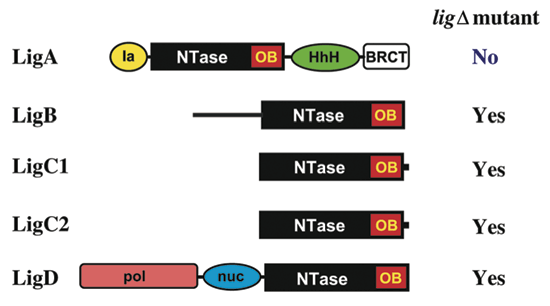
of the four DNA ligases of mycobacteria. _J. Biol. Chem._ 279, 20594–20606 (2004). Article CAS Google Scholar * Aravind, L. & Koonin, E.V. Prokaryotic homologs of the eukaryotic
DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. _Genome Res._ 11, 1365–1374 (2001). Article CAS Google
Scholar * Doherty, A.J., Jackson, S.P. & Weller, G.R. Identification of bacterial homologues of the Ku DNA repair proteins. _FEBS Lett._ 500, 186–188 (2001). Article CAS Google
Scholar * Della, M. et al. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. _Science_ 306, 683–685 (2004). Article CAS Google Scholar * Zhu, H. &
Shuman, S. A primer-dependent polymerase function of _Pseudomonas aeruginosa_ ATP-dependent DNA ligase (LigD). _J. Biol. Chem._ 280, 418–427 (2005). Article CAS Google Scholar * Weller,
G.R. et al. Identification of a DNA nonhomologous end-joining complex in bacteria. _Science_ 297, 1686–1689 (2002). Article CAS Google Scholar * Braunstein, M., Brown, A.M., Kurtz, S.
& Jacobs, W.R. Jr. Two nonredundant SecA homologues function in mycobacteria. _J. Bacteriol._ 183, 6979–6990 (2001). Article CAS Google Scholar * Lipps, G., Weinzierl, A.O., von
Scheven, G., Buchen, C. & Cramer, P. Structure of a bifunctional DNA primase-polymerase. _Nat. Struct. Mol. Biol._ 11, 157–162 (2004). Article CAS Google Scholar * Ito, N., Nureki,
O., Shirouzu, M., Yokoyama, S. & Hanaoka, F. Crystal structure of the _Pyrococcus horikoshii_ DNA primase-UTP complex: implications for the mechanism of primer synthesis. _Genes Cells_
8, 913–923 (2003). Article CAS Google Scholar * Augustin, M.A., Huber, R. & Kaiser, J.T. Crystal structure of a DNA-dependent RNA polymerase (DNA primase). _Nat. Struct. Biol._ 8,
57–61 (2001). Article CAS Google Scholar * Glickman, M.S. The _mmaA2_ gene of _Mycobacterium tuberculosis_ encodes the distal cyclopropane synthase of the α-mycolic acid. _J. Biol. Chem._
278, 7844–7849 (2003). Article CAS Google Scholar * Manolis, K.G. et al. Novel functional requirements for non-homologous DNA end joining in _Schizosaccharomyces pombe_. _EMBO J._ 20,
210–221 (2001). Article CAS Google Scholar * Walker, J.R., Corpina, R.A. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break
repair. _Nature_ 412, 607–614 (2001). Article CAS Google Scholar * Aylon, Y., Liefshitz, B. & Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination
during the cell cycle. _EMBO J._ 23, 4868–4875 (2004). Article CAS Google Scholar * Heidenreich, E., Novotny, R., Kneidinger, B., Holzmann, V. & Wintersberger, U. Non-homologous end
joining as an important mutagenic process in cell cycle– arrested cells. _EMBO J._ 22, 2274–2283 (2003). Article CAS Google Scholar * Ferreira, M.G. & Cooper, J.P. Two modes of DNA
double-strand break repair are reciprocally regulated through the fission yeast cell cycle. _Genes Dev._ 18, 2249–2254 (2004). Article CAS Google Scholar * Timm, J., Lim, E.M. &
Gicquel, B. _Escherichia coli_– mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. _J. Bacteriol._ 176, 6749–6753 (1994). Article CAS Google Scholar *
Golemis, E. et al. The interaction trap. In _Current Protocols in Molecular Biology_ (eds. Ausubel, F.M. et al.) 20.1.1–20.1.2 (Wiley, New York, 1999). Google Scholar Download references
ACKNOWLEDGEMENTS This research was supported by US National Institutes of Health grants AI53417 (to M.S.G.) and GM63611 (to S.S.). M.S.G. is the recipient of research awards from the Ellison
Medical Foundation and the New York Academy of Medicine Speakers Fund for Biomedical Research. S.S. is an American Cancer Society Research Professor. AUTHOR INFORMATION Author notes *
Chunling Gong and Paola Bongiorno: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Immunology and Molecular Biology Programs, Sloan-Kettering Institute, New York,
10021, New York, USA Chunling Gong, Paola Bongiorno, Alexandra Martins, Nicolas C Stephanou, Hui Zhu, Stewart Shuman & Michael S Glickman * Division of Infectious Diseases, Memorial
Sloan Kettering Cancer Center, New York, 10021, New York, USA Chunling Gong, Paola Bongiorno, Alexandra Martins, Nicolas C Stephanou, Hui Zhu, Stewart Shuman & Michael S Glickman Authors
* Chunling Gong View author publications You can also search for this author inPubMed Google Scholar * Paola Bongiorno View author publications You can also search for this author inPubMed
Google Scholar * Alexandra Martins View author publications You can also search for this author inPubMed Google Scholar * Nicolas C Stephanou View author publications You can also search for
this author inPubMed Google Scholar * Hui Zhu View author publications You can also search for this author inPubMed Google Scholar * Stewart Shuman View author publications You can also
search for this author inPubMed Google Scholar * Michael S Glickman View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence
to Stewart Shuman or Michael S Glickman. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1
_Mycobacterium smegmatis_ deletion strains. (PDF 19 kb) SUPPLEMENTARY METHODS (PDF 89 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gong, C.,
Bongiorno, P., Martins, A. _et al._ Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. _Nat Struct Mol Biol_ 12,
304–312 (2005). https://doi.org/10.1038/nsmb915 Download citation * Received: 03 February 2005 * Accepted: 01 March 2005 * Published: 20 March 2005 * Issue Date: 01 April 2005 * DOI:
https://doi.org/10.1038/nsmb915 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative