- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
KEY POINTS * Alternative polyadenylation (APA) is a widespread mechanism of gene regulation that generates distinct 3′ ends in transcripts made by RNA polymerase II. * APA is tissue specific
and globally regulated in various conditions, such as cell proliferation and differentiation, and in response to extracellular cues. * APA occurring in 3′ untranslated regions (3′ UTRs)
leads to the production of mRNA isoforms with different metabolisms and can also affect protein localization. * APA occurring in the region upstream of the 3′ UTR is often coupled with
splicing and can lead to the production of distinct protein isoforms. It can also function by repressing gene expression. * APA is regulated by several known mechanisms, including regulation
of the levels of core RNA-processing factors and other RNA-binding proteins, as well as by splicing and transcriptional dynamics. ABSTRACT Alternative polyadenylation (APA) is an
RNA-processing mechanism that generates distinct 3′ termini on mRNAs and other RNA polymerase II transcripts. It is widespread across all eukaryotic species and is recognized as a major
mechanism of gene regulation. APA exhibits tissue specificity and is important for cell proliferation and differentiation. In this Review, we discuss the roles of APA in diverse cellular
processes, including mRNA metabolism, protein diversification and protein localization, and more generally in gene regulation. We also discuss the molecular mechanisms underlying APA, such
as variation in the concentration of core processing factors and RNA-binding proteins, as well as transcription-based regulation. Access through your institution Buy or subscribe This is a
preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per
year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS ALTERNATIVE
POLYADENYLATION BY SEQUENTIAL ACTIVATION OF DISTAL AND PROXIMAL POLYA SITES Article 10 January 2022 CONTEXT-SPECIFIC REGULATION AND FUNCTION OF MRNA ALTERNATIVE POLYADENYLATION Article 07
July 2022 EXTENSIVE 5′-SURVEILLANCE GUARDS AGAINST NON-CANONICAL NAD-CAPS OF NUCLEAR MRNAS IN YEAST Article Open access 02 November 2020 REFERENCES * Richard, P. & Manley, J. L.
Transcription termination by nuclear RNA polymerases. _Genes Dev._ 23, 1247–1269 (2009). Article CAS PubMed PubMed Central Google Scholar * Marzluff, W. F., Wagner, E. J. & Duronio,
R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. _Nat. Rev. Genet._ 9, 843–854 (2008). Article CAS PubMed PubMed Central Google Scholar * Tian,
B. & Graber, J. H. Signals for pre-mRNA cleavage and polyadenylation. _Wiley Interdiscip. Rev. RNA_ 3, 385–396 (2012). Article CAS PubMed Google Scholar * Mandel, C. R., Bai, Y.
& Tong, L. Protein factors in pre-mRNA 3′-end processing. _Cell. Mol. Life Sci._ 65, 1099–1122 (2008). Article CAS PubMed PubMed Central Google Scholar * Zhao, J., Hyman, L. &
Moore, C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. _Microbiol. Mol. Biol. Rev._ 63, 405–445 (1999). Article
CAS PubMed PubMed Central Google Scholar * Shi, Y. & Manley, J. L. The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. _Genes Dev._ 29,
889–897 (2015). Article CAS PubMed PubMed Central Google Scholar * Colgan, D. F. & Manley, J. L. Mechanism and regulation of mRNA polyadenylation. _Genes Dev._ 11, 2755–2766 (1997).
Article CAS PubMed Google Scholar * Edwalds-Gilbert, G., Veraldi, K. L. & Milcarek, C. Alternative poly(A) site selection in complex transcription units: means to an end? _Nucleic
Acids Res._ 25, 2547–2561 (1997). Article CAS PubMed PubMed Central Google Scholar * Barabino, S. M. & Keller, W. Last but not least: regulated poly(A) tail formation. _Cell_ 99,
9–11 (1999). Article CAS PubMed Google Scholar * Gautheret, D., Poirot, O., Lopez, F., Audic, S. & Claverie, J. M. Alternate polyadenylation in human mRNAs: a large-scale analysis by
EST clustering. _Genome Res._ 8, 524–530 (1998). THIS PAPER REPORTS THE FIRST USE OF EXPRESSED SEQUENCE TAGS TO IDENTIFY APA SITES GENOME-WIDE. Article CAS PubMed Google Scholar * Tian,
B., Hu, J., Zhang, H. & Lutz, C. S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. _Nucleic Acids Res._ 33, 201–212 (2005). CAS PubMed PubMed Central Google
Scholar * Derti, A. et al. A quantitative atlas of polyadenylation in five mammals. _Genome Res._ 22, 1173–1183 (2012). Article CAS PubMed PubMed Central Google Scholar * Hoque, M. et
al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. _Nat. Methods_ 10, 133–139 (2013). Article CAS PubMed Google Scholar * Mayr, C.
Evolution and biological roles of alternative 3′ UTRs. _Trends Cell Biol._ 26, 227–237 (2016). Article CAS PubMed Google Scholar * Proudfoot, N. J. Ending the message: poly(A) signals
then and now. _Genes Dev._ 25, 1770–1782 (2011). Article CAS PubMed PubMed Central Google Scholar * Elkon, R., Ugalde, A. P. & Agami, R. Alternative cleavage and polyadenylation:
extent, regulation and function. _Nat. Rev. Genet._ 14, 496–506 (2013). Article CAS PubMed Google Scholar * Di Giammartino, D. C., Nishida, K. & Manley, J. L. Mechanisms and
consequences of alternative polyadenylation. _Mol. Cell_ 43, 853–866 (2011). Article CAS PubMed PubMed Central Google Scholar * Tian, B. & Manley, J. L. Alternative cleavage and
polyadenylation: the long and short of it. _Trends Biochem. Sci._ 38, 312–320 (2013). Article CAS PubMed PubMed Central Google Scholar * Hunt, A. G. Messenger RNA 3′ end formation in
plants. _Curr. Top. Microbiol. Immunol._ 326, 151–177 (2008). CAS PubMed Google Scholar * Bartel, D. P. MicroRNAs: target recognition and regulatory functions. _Cell_ 136, 215–233 (2009).
Article CAS PubMed PubMed Central Google Scholar * Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A. & Burge, C. B. Proliferating cells express mRNAs with shortened 3′
untranslated regions and fewer microRNA target sites. _Science_ 320, 1643–1647 (2008). THIS IS THE FIRST REPORT THAT GLOBAL CHANGES IN APA OCCUR AS A CONSEQUENCE OF CHANGES IN CELL
PROLIFERATION, SPECIFICALLY DEMONSTRATING THE USE OF PROXIMAL 3′ UTR PASS DURING THE ACTIVATION OF T CELLS. Article CAS PubMed PubMed Central Google Scholar * Ji, Z., Lee, J. Y., Pan,
Z., Jiang, B. & Tian, B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. _Proc. Natl Acad. Sci. USA_ 106,
7028–7033 (2009). THIS ARTICLE DESCRIBES GLOBAL APA REGULATION IN EMBRYONIC DEVELOPMENT, CONNECTING POLYADENYLATION ACTIVITY WITH APA DURING CELL DIFFERENTIATION. Article CAS PubMed
PubMed Central Google Scholar * Mayr, C. & Bartel, D. P. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. _Cell_ 138,
673–684 (2009). THIS PUBLICATION REPORTS A CONNECTION BETWEEN 3′ UTR SHORTENING, ESPECIALLY IN THE TRANSCRIPTS OF SEVERAL PROTO-ONCOGENES, AND CELL TRANSFORMATION. Article CAS PubMed
PubMed Central Google Scholar * Nam, J. W. et al. Global analyses of the effect of different cellular contexts on microRNA targeting. _Mol. Cell_ 53, 1031–1043 (2014). Article CAS PubMed
PubMed Central Google Scholar * Hoffman, Y. et al. 3′ UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. _PLoS Genet._ 12,
e1005879 (2016). Article PubMed PubMed Central CAS Google Scholar * Garneau, N. L., Wilusz, J. & Wilusz, C. J. The highways and byways of mRNA decay. _Nat. Rev. Mol. Cell Biol._ 8,
113–126 (2007). Article CAS PubMed Google Scholar * Graham, R. R. et al. Three functional variants of IFN regulatory factor 5 (_IRF5_) define risk and protective haplotypes for human
lupus. _Proc. Natl Acad. Sci. USA_ 104, 6758–6763 (2007). Article CAS PubMed PubMed Central Google Scholar * Gong, C. & Maquat, L. E. lncRNAs transactivate STAU1-mediated mRNA decay
by duplexing with 3′ UTRs via Alu elements. _Nature_ 470, 284–288 (2011). Article CAS PubMed PubMed Central Google Scholar * Hogg, J. R. & Goff, S. P. Upf1 senses 3′ UTR length to
potentiate mRNA decay. _Cell_ 143, 379–389 (2010). Article CAS PubMed PubMed Central Google Scholar * Spies, N., Burge, C. B. & Bartel, D. P. 3′ UTR-isoform choice has limited
influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. _Genome Res._ 23, 2078–2090 (2013). THIS REPORT PRESENTS A GLOBAL ANALYSIS OF THE DIFFERENT
EFFECTS OF SHORT AND LONG 3′ UTRS ON MRNA DECAY AND TRANSLATION. Article CAS PubMed PubMed Central Google Scholar * Ulitsky, I. et al. Extensive alternative polyadenylation during
zebrafish development. _Genome Res._ 22, 2054–2066 (2012). Article CAS PubMed PubMed Central Google Scholar * Geisberg, J. V., Moqtaderi, Z., Fan, X., Ozsolak, F. & Struhl, K.
Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. _Cell_ 156, 812–824 (2014). Article CAS PubMed PubMed Central Google Scholar *
Tycowski, K. T., Shu, M. D. & Steitz, J. A. Myriad triple-helix-forming structures in the transposable element RNAs of plants and fungi. _Cell Rep._ 15, 1266–1276 (2016). Article CAS
PubMed PubMed Central Google Scholar * Lee, J. E., Lee, J. Y., Wilusz, J., Tian, B. & Wilusz, C. J. Systematic analysis of _cis_-elements in unstable mRNAs demonstrates that CUGBP1 is
a key regulator of mRNA decay in muscle cells. _PLoS ONE_ 5, e11201 (2010). Article PubMed PubMed Central CAS Google Scholar * Floor, S. N. & Doudna, J. A. Tunable protein
synthesis by transcript isoforms in human cells. _eLife_ 5, e10921 (2016). Article PubMed PubMed Central Google Scholar * Neve, J. et al. Subcellular RNA profiling links splicing and
nuclear DICER1 to alternative cleavage and polyadenylation. _Genome Res._ 26, 24–35 (2016). Article CAS PubMed PubMed Central Google Scholar * Djebali, S. et al. Landscape of
transcription in human cells. _Nature_ 489, 101–108 (2012). Article CAS PubMed PubMed Central Google Scholar * Chen, L. L. & Carmichael, G. G. Altered nuclear retention of mRNAs
containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. _Mol. Cell_ 35, 467–478 (2009). Article CAS PubMed PubMed Central Google Scholar *
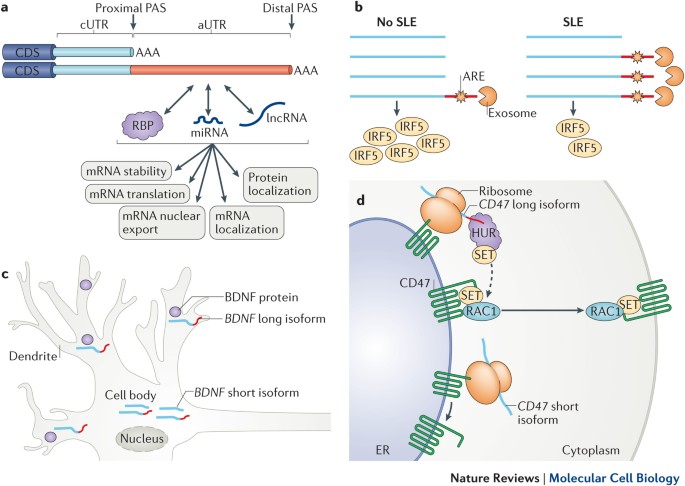
Martin, K. C. & Ephrussi, A. mRNA localization: gene expression in the spatial dimension. _Cell_ 136, 719–730 (2009). Article CAS PubMed PubMed Central Google Scholar * An, J. J.
et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. _Cell_ 134, 175–187 (2008). Article CAS PubMed PubMed Central Google
Scholar * Andreassi, C. & Riccio, A. To localize or not to localize: mRNA fate is in 3′ UTR ends. _Trends Cell Biol._ 19, 465–474 (2009). Article CAS PubMed Google Scholar * Yudin,
D. et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. _Neuron_ 59, 241–252 (2008). Article CAS PubMed PubMed Central Google Scholar
* Taliaferro, J. M. et al. Distal alternative last exons localize mRNAs to neural projections. _Mol. Cell_ 61, 821–833 (2016). Article CAS PubMed PubMed Central Google Scholar * Loya,
A. et al. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. _RNA_ 14, 1352–1365 (2008). Article CAS PubMed PubMed Central Google Scholar
* Reid, D. W. & Nicchitta, C. V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. _Nat. Rev. Mol. Cell Biol._ 16, 221–231 (2015). Article CAS PubMed
PubMed Central Google Scholar * Berkovits, B. D. & Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. _Nature_ 522, 363–367 (2015). THIS WORK
DISCOVERS A NOVEL MECHANISM BY WHICH THE AUTR OF A TRANSCRIPT FUNCTIONS AS A SCAFFOLD FOR THE ASSEMBLY OF SPECIFIC PROTEIN COMPLEXES, WHICH THEN MODULATE THE SUBCELLULAR LOCALIZATION OF THE
ENCODED PROTEIN. Article CAS PubMed PubMed Central Google Scholar * Vasudevan, S., Peltz, S. W. & Wilusz, C. J. Non-stop decay — a new mRNA surveillance pathway. _Bioessays_ 24,
785–788 (2002). Article CAS PubMed Google Scholar * Yao, P. et al. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. _Cell_ 149,
88–100 (2012). Article CAS PubMed PubMed Central Google Scholar * Elkon, R. et al. E2F mediates enhanced alternative polyadenylation in proliferation. _Genome Biol._ 13, R59 (2012).
Article CAS PubMed PubMed Central Google Scholar * Amara, S. G., Jonas, V., Rosenfeld, M. G., Ong, E. S. & Evans, R. M. Alternative RNA processing in calcitonin gene expression
generates mRNAs encoding different polypeptide products. _Nature_ 298, 240–244 (1982). Article CAS PubMed Google Scholar * Alt, F. W. et al. Synthesis of secreted and membrane-bound
immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. _Cell_ 20, 293–301 (1980). Article CAS PubMed Google Scholar * Davis, M. J. et al. Differential use of
signal peptides and membrane domains is a common occurrence in the protein output of transcriptional units. _PLoS Genet._ 2, e46 (2006). Article PubMed PubMed Central CAS Google Scholar
* Vorlova, S. et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. _Mol. Cell_ 43, 927–939 (2011). Article CAS PubMed PubMed Central
Google Scholar * Di Giammartino, D. C. et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. _Gene Dev._ 28, 2248–2260
(2014). Article PubMed CAS PubMed Central Google Scholar * Mbita, Z. et al. De-regulation of the RBBP6 isoform 3/DWNN in human cancers. _Mol. Cell Biochem._ 362, 249–262 (2012). Article
CAS PubMed Google Scholar * Pan, Z. et al. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. _Gene_ 366,
325–334 (2006). Article CAS PubMed Google Scholar * Luo, W. et al. The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity
through feedback autoregulation and by U1 snRNP. _PLoS Genet._ 9, e1003613 (2013). Article CAS PubMed PubMed Central Google Scholar * Audibert, A. & Simonelig, M. Autoregulation at
the level of mRNA 3′ end formation of the _suppressor of forked_ gene of _Drosophila melanogaster_ is conserved in _Drosophila virilis_. _Proc. Natl Acad. Sci. USA_ 95, 14302–14307 (1998).
Article CAS PubMed PubMed Central Google Scholar * Zhao, W. & Manley, J. L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms.
_Mol. Cell. Biol._ 16, 2378–2386 (1996). Article CAS PubMed PubMed Central Google Scholar * Takagaki, Y., Seipelt, R. L., Peterson, M. L. & Manley, J. L. The polyadenylation factor
CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. _Cell_ 87, 941–952 (1996). THIS STUDY UNCOVERS A MECHANISM OF APA REGULATION IN WHICH
INCREASED EXPRESSION OF A CORE POLYADENYLATION FACTOR, CSTF64, DURING B CELL DIFFERENTIATION SHIFTS PAS USAGE TO AN UPSTREAM SITE IN THE IGM HEAVY CHAIN PRE-MRNA. Article CAS PubMed
Google Scholar * Yao, C. et al. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3′ processing. _RNA_ 19, 1781–1790 (2013). Article CAS PubMed PubMed Central
Google Scholar * Li, W. et al. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and
polyadenylation. _PLoS Genet._ 11, e1005166 (2015). Article PubMed PubMed Central CAS Google Scholar * Xia, Z. et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal
a 3′-UTR landscape across seven tumour types. _Nat. Commun._ 5, 5274 (2014). Article CAS PubMed Google Scholar * Ji, Z. & Tian, B. Reprogramming of 3′ untranslated regions of mRNAs
by alternative polyadenylation in generation of pluripotent stem cells from different cell types. _PLoS ONE_ 4, e8419 (2009). Article PubMed PubMed Central CAS Google Scholar *
Lackford, B. et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. _EMBO J._ 33, 878–889 (2014). Article CAS PubMed PubMed Central Google Scholar *
Martin, G., Gruber, A. R., Keller, W. & Zavolan, M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR
length. _Cell Rep._ 1, 753–763 (2012). Article CAS PubMed Google Scholar * Gruber, A. R., Martin, G., Keller, W. & Zavolan, M. Cleavage factor Im is a key regulator of 3′ UTR length.
_RNA Biol._ 9, 1405–1412 (2012). Article CAS PubMed Google Scholar * Brown, K. M. & Gilmartin, G. M. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage
factor Im . _Mol. Cell_ 12, 1467–1476 (2003). Article CAS PubMed Google Scholar * Yang, Q., Gilmartin, G. M. & Doublié, S. The structure of human Cleavage Factor Im hints at
functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. _RNA Biol._ 8, 748–753 (2011). Article CAS PubMed PubMed
Central Google Scholar * Masamha, C. P. et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. _Nature_ 510, 412–416 (2014). Article CAS PubMed PubMed
Central Google Scholar * Gennarino, V. A. et al. _NUDT21_-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. _eLife_ 4, e10782
(2015). Article PubMed Central Google Scholar * Kuhn, U. et al. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A)
polymerase and the cleavage and polyadenylation specificity factor. _J. Biol. Chem._ 284, 22803–22814 (2009). Article PubMed PubMed Central CAS Google Scholar * Jenal, M. et al. The
poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. _Cell_ 149, 538–553 (2012). Article CAS PubMed Google Scholar * de Klerk, E. et al. Poly(A)
binding protein nuclear 1 levels affect alternative polyadenylation. _Nucleic Acids Res._ 40, 9089–9101 (2012). Article PubMed PubMed Central CAS Google Scholar * Bresson, S. M. &
Conrad, N. K. The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. _PLoS Genet._ 9, e1003893 (2013). Article PubMed PubMed Central CAS Google Scholar *
Beaulieu, Y. B., Kleinman, C. L., Landry-Voyer, A. M., Majewski, J. & Bachand, F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein
nuclear 1. _PLoS Genet._ 8, e1003078 (2012). Article CAS PubMed PubMed Central Google Scholar * Bresson, S. M., Hunter, O. V., Hunter, A. C. & Conrad, N. K. Canonical poly(A)
polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. _PLoS Genet._ 11, e1005610 (2015). Article PubMed PubMed Central CAS Google Scholar * Thomas, P. E.
et al. Genome-wide control of polyadenylation site choice by CPSF30 in _Arabidopsis_. _Plant Cell_ 24, 4376–4388 (2012). Article CAS PubMed PubMed Central Google Scholar * Niwa, M.,
Rose, S. D. & Berget, S. M. _In vitro_ polyadenylation is stimulated by the presence of an upstream intron. _Genes Dev._ 4, 1552–1559 (1990). Article CAS PubMed Google Scholar *
Tian, B., Pan, Z. & Lee, J. Y. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. _Genome Res._ 17, 156–165 (2007).
Article CAS PubMed PubMed Central Google Scholar * Lutz, C. S. et al. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor
increases polyadenylation efficiency _in vitro_. _Genes Dev._ 10, 325–337 (1996). Article CAS PubMed Google Scholar * Kyburz, A., Friedlein, A., Langen, H. & Keller, W. Direct
interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. _Mol. Cell_ 23, 195–205 (2006). Article CAS PubMed Google
Scholar * Millevoi, S. et al. An interaction between U2AF 65 and CF Im links the splicing and 3′ end processing machineries. _EMBO J._ 25, 4854–4864 (2006). Article CAS PubMed PubMed
Central Google Scholar * Gunderson, S. I., Polycarpou-Schwarz, M. & Mattaj, I. W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A)
polymerase. _Mol. Cell_ 1, 255–264 (1998). Article CAS PubMed Google Scholar * Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. _Nature_ 468,
664–668 (2010). THIS ARTICLE DESCRIBES A GLOBAL ACTIVITY OF U1 SNRNP IN SUPPRESSING PROMOTER-PROXIMAL PASS. Article CAS PubMed PubMed Central Google Scholar * Berg, M. G. et al. U1
snRNP determines mRNA length and regulates isoform expression. _Cell_ 150, 53–64 (2012). Article CAS PubMed PubMed Central Google Scholar * Engreitz, J. M. et al. RNA−RNA interactions
enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. _Cell_ 159, 188–199 (2014). Article CAS PubMed PubMed Central Google Scholar * Wahl, M. C., Will,
C. L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. _Cell_ 136, 701–718 (2009). Article CAS PubMed Google Scholar * Devany, E. et al. Intronic cleavage
and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. _Cell Discov._ 2, 16013 (2016). Article CAS PubMed PubMed Central Google Scholar * Licatalosi,
D. D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. _Nature_ 456, 464–469 (2008). THIS IS THE FIRST DEMONSTRATION THAT A SPLICING-REGULATORY RBP, NOVA,
CAN ALSO REGULATE APA. Article CAS PubMed PubMed Central Google Scholar * Zheng, D. & Tian, B. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. _Adv.
Exp. Med. Biol._ 825, 97–127 (2014). Article CAS PubMed Google Scholar * Hilgers, V., Lemke, S. B. & Levine, M. ELAV mediates 3′ UTR extension in the _Drosophila_ nervous system.
_Genes Dev._ 26, 2259–2264 (2012). Article CAS PubMed PubMed Central Google Scholar * Oktaba, K. et al. ELAV links paused Pol II to alternative polyadenylation in the _Drosophila_
nervous system. _Mol. Cell_ 57, 341–348 (2015). Article CAS PubMed Google Scholar * Zhu, H., Zhou, H. L., Hasman, R. A. & Lou, H. Hu proteins regulate polyadenylation by blocking
sites containing U-rich sequences. _J. Biol. Chem._ 282, 2203–2210 (2007). Article CAS PubMed Google Scholar * Dai, W., Zhang, G. & Makeyev, E. V. RNA-binding protein HuR
autoregulates its expression by promoting alternative polyadenylation site usage. _Nucleic Acids Res._ 40, 787–800 (2012). Article CAS PubMed Google Scholar * Mansfield, K. D. &
Keene, J. D. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. _Nucleic Acids Res._ 40, 2734–2746 (2012). Article CAS
PubMed Google Scholar * Manley, J. L. & Tacke, R. SR proteins and splicing control. _Genes Dev._ 10, 1569–1579 (1996). Article CAS PubMed Google Scholar * Howard, J. M. &
Sanford, J. R. The RNAissance family: SR proteins as multifaceted regulators of gene expression. _Wiley Interdiscip. Rev. RNA_ 6, 93–110 (2015). Article CAS PubMed Google Scholar *
Muller-McNicoll, M. et al. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. _Genes Dev._ 30, 553–566 (2016). Article PubMed PubMed Central Google Scholar
* Tran, D. D. et al. THOC5 controls 3′ end-processing of immediate early genes via interaction with polyadenylation specific factor 100 (CPSF100). _Nucleic Acids Res._ 42, 12249–12260
(2014). Article CAS PubMed PubMed Central Google Scholar * Johnson, S. A., Kim, H., Erickson, B. & Bentley, D. L. The export factor Yra1 modulates mRNA 3′ end processing. _Nat.
Struct. Mol. Biol._ 18, 1164–1171 (2011). Article CAS PubMed PubMed Central Google Scholar * Ling, S. C., Polymenidou, M. & Cleveland, D. W. Converging mechanisms in ALS and FTD:
disrupted RNA and protein homeostasis. _Neuron_ 79, 416–438 (2013). Article CAS PubMed PubMed Central Google Scholar * Masuda, A. et al. Position-specific binding of FUS to nascent RNA
regulates mRNA length. _Genes Dev._ 29, 1045–1057 (2015). Article CAS PubMed PubMed Central Google Scholar * Schwartz, J. C. et al. FUS binds the CTD of RNA polymerase II and regulates
its phosphorylation at Ser2. _Genes Dev._ 26, 2690–2695 (2012). Article CAS PubMed PubMed Central Google Scholar * Hoell, J. I. et al. RNA targets of wild-type and mutant FET family
proteins. _Nat. Struct. Mol. Biol._ 18, 1428–1431 (2011). Article CAS PubMed PubMed Central Google Scholar * Prudencio, M. et al. Distinct brain transcriptome profiles in
_C9orf72_-associated and sporadic ALS. _Nat. Neurosci._ 18, 1175–1182 (2015). Article CAS PubMed PubMed Central Google Scholar * Lee, Y. B. et al. Hexanucleotide repeats in ALS/FTD form
length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. _Cell Rep._ 5, 1178–1186 (2013). Article CAS PubMed PubMed Central Google Scholar * Batra, R. et al. Loss
of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. _Mol. Cell_ 56, 311–322 (2014). Article CAS PubMed PubMed Central Google
Scholar * Naftelberg, S., Schor, I. E., Ast, G. & Kornblihtt, A. R. Regulation of alternative splicing through coupling with transcription and chromatin structure. _Annu. Rev. Biochem._
84, 165–198 (2015). Article CAS PubMed Google Scholar * Yonaha, M. & Proudfoot, N. J. Specific transcriptional pausing activates polyadenylation in a coupled _in vitro_ system.
_Mol. Cell_ 3, 593–600 (1999). Article CAS PubMed Google Scholar * Cui, Y. & Denis, C. L. _In vivo_ evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and
SPT5 enhance upstream poly(A) site utilization. _Mol. Cell. Biol._ 23, 7887–7901 (2003). Article CAS PubMed PubMed Central Google Scholar * Martincic, K., Alkan, S. A., Cheatle, A.,
Borghesi, L. & Milcarek, C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. _Nat. Immunol._ 10, 1102–1109
(2009). Article CAS PubMed PubMed Central Google Scholar * Pinto, P. A. et al. RNA polymerase II kinetics in _polo_ polyadenylation signal selection. _EMBO J._ 30, 2431–2444 (2011).
Article CAS PubMed PubMed Central Google Scholar * Rosonina, E., Bakowski, M. A., McCracken, S. & Blencowe, B. J. Transcriptional activators control splicing and 3′ -end cleavage
levels. _J. Biol. Chem._ 278, 43034–43040 (2003). Article CAS PubMed Google Scholar * Nagaike, T. et al. Transcriptional activators enhance polyadenylation of mRNA precursors. _Mol.
Cell_ 41, 409–418 (2011). Article CAS PubMed PubMed Central Google Scholar * Ji, Z. et al. Transcriptional activity regulates alternative cleavage and polyadenylation. _Mol. Syst.
Biol._ 7, 534 (2011). Article PubMed PubMed Central CAS Google Scholar * Ni, T. et al. Distinct polyadenylation landscapes of diverse human tissues revealed by a modified PA-seq
strategy. _BMC Genomics_ 14, 615 (2013). Article CAS PubMed PubMed Central Google Scholar * Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D. L. RNA polymerase II pauses and
associates with pre-mRNA processing factors at both ends of genes. _Nat. Struct. Mol. Biol._ 15, 71–78 (2008). Article CAS PubMed Google Scholar * Venkataraman, K., Brown, K. M. &
Gilmartin, G. M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. _Genes Dev._ 19, 1315–1327 (2005). Article CAS PubMed
PubMed Central Google Scholar * Rozenblatt-Rosen, O. et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. _Proc. Natl Acad. Sci. USA_
106, 755–760 (2009). Article CAS PubMed PubMed Central Google Scholar * Calvo, O. & Manley, J. L. Strange bedfellows: polyadenylation factors at the promoter. _Genes Dev._ 17,
1321–1327 (2003). Article CAS PubMed Google Scholar * Uhlmann, T., Boeing, S., Lehmbacher, M. & Meisterernst, M. The VP16 activation domain establishes an active mediator lacking
CDK8 _in vivo_. _J. Biol. Chem._ 282, 2163–2173 (2007). Article CAS PubMed Google Scholar * Yang, Y. et al. PAF complex plays novel subunit-specific roles in alternative cleavage and
polyadenylation. _PLoS Genet._ 12, e1005794 (2016). Article PubMed PubMed Central CAS Google Scholar * Yu, M. et al. RNA polymerase II-associated factor 1 regulates the release and
phosphorylation of paused RNA polymerase II. _Science_ 350, 1383–1386 (2015). Article CAS PubMed PubMed Central Google Scholar * Jiang, C. & Pugh, B. F. Nucleosome positioning and
gene regulation: advances through genomics. _Nat. Rev. Genet._ 10, 161–172 (2009). Article CAS PubMed PubMed Central Google Scholar * Kaplan, N. et al. The DNA-encoded nucleosome
organization of a eukaryotic genome. _Nature_ 458, 362–366 (2009). Article CAS PubMed Google Scholar * Spies, N., Nielsen, C. B., Padgett, R. A. & Burge, C. B. Biased chromatin
signatures around polyadenylation sites and exons. _Mol. Cell_ 36, 245–254 (2009). Article CAS PubMed PubMed Central Google Scholar * Grosso, A. R., de Almeida, S. F., Braga, J. &
Carmo-Fonseca, M. Dynamic transitions in RNA polymerase II density profiles during transcription termination. _Genome Res._ 22, 1447–1456 (2012). Article CAS PubMed PubMed Central Google
Scholar * Li, W. et al. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. _BMC Biol._ 14, 6 (2016). Article
PubMed PubMed Central CAS Google Scholar * Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. _Genes Dev._ 29, 2037–2053
(2015). Article CAS PubMed PubMed Central Google Scholar * Eckmann, C. R., Rammelt, C. & Wahle, E. Control of poly(A) tail length. _Wiley Interdiscip. Rev. RNA_ 2, 348–361 (2011).
Article CAS PubMed Google Scholar * Schmidt, M. J. & Norbury, C. J. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. _Wiley Interdiscip. Rev. RNA_ 1,
142–151 (2010). Article CAS PubMed Google Scholar * Mendez, R. & Richter, J. D. Translational control by CPEB: a means to the end. _Nat. Rev. Mol. Cell Biol._ 2, 521–529 (2001).
Article CAS PubMed Google Scholar * Subtelny, A. O., Eichhorn, S. W., Chen, G. R., Sive, H. & Bartel, D. P. Poly(A)-tail profiling reveals an embryonic switch in translational
control. _Nature_ 508, 66–71 (2014). Article CAS PubMed PubMed Central Google Scholar * Chang, H., Lim, J., Ha, M. & Kim, V. N. TAIL-seq: genome-wide determination of poly(A) tail
length and 3′ end modifications. _Mol. Cell_ 53, 1044–1052 (2014). Article CAS PubMed Google Scholar * Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. _Mol.
Cell_ 43, 904–914 (2011). Article CAS PubMed PubMed Central Google Scholar * Naganuma, T. et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear
paraspeckles. _EMBO J._ 31, 4020–4034 (2012). Article CAS PubMed PubMed Central Google Scholar * Higgs, D. R. et al. α-Thalassaemia caused by a polyadenylation signal mutation. _Nature_
306, 398–400 (1983). Article CAS PubMed Google Scholar * Prasad, M. K. et al. A polymorphic 3′ UTR element in ATP1B1 regulates alternative polyadenylation and is associated with blood
pressure. _PLoS ONE_ 8, e76290 (2013). Article CAS PubMed PubMed Central Google Scholar * Singh, P. et al. Global changes in processing of mRNA 3′ untranslated regions characterize
clinically distinct cancer subtypes. _Cancer Res._ 69, 9422–9430 (2009). Article CAS PubMed PubMed Central Google Scholar * Creemers, E. E. et al. Genome-wide polyadenylation maps
reveal dynamic mRNA 3′-end formation in the failing human heart. _Circ. Res._ 118, 433–438 (2016). Article CAS PubMed Google Scholar * Soetanto, R. et al. Role of miRNAs and alternative
mRNA 3′-end cleavage and polyadenylation of their mRNA targets in cardiomyocyte hypertrophy. _Biochim. Biophys. Acta_ 1859, 744–756 (2016). Article CAS PubMed Google Scholar * Park, J.
Y. et al. Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. _PLoS ONE_ 6, e22391 (2011).
Article CAS PubMed PubMed Central Google Scholar * Hu, J., Lutz, C. S., Wilusz, J. & Tian, B. Bioinformatic identification of candidate _cis_-regulatory elements involved in human
mRNA polyadenylation. _RNA_ 11, 1485–1493 (2005). Article CAS PubMed PubMed Central Google Scholar * Cheng, Y., Miura, R. M. & Tian, B. Prediction of mRNA polyadenylation sites by
support vector machine. _Bioinformatics_ 22, 2320–2325 (2006). Article CAS PubMed Google Scholar * Nunes, N. M., Li, W., Tian, B. & Furger, A. A functional human poly(A) site
requires only a potent DSE and an A-rich upstream sequence. _EMBO J._ 29, 1523–1536 (2010). Article CAS PubMed PubMed Central Google Scholar * Sheets, M. D., Ogg, S. C. & Wickens,
M. P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation _in vitro_. _Nucleic Acids Res._ 18, 5799–5805 (1990).
Article CAS PubMed PubMed Central Google Scholar * Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. _Mol. Cell_ 33, 365–376 (2009). THIS REPORT DETAILS
THE PURIFICATION OF AN ACTIVE POLYADENYLATION COMPLEX ON SUBSTRATE RNA AND THE IDENTIFICATION OF MORE THAN 80 CORE AND ASSOCIATED PROTEINS. Article CAS PubMed PubMed Central Google
Scholar * Chan, S. L. et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. _Genes Dev._ 28, 2370–2380 (2014). Article PubMed PubMed Central CAS Google
Scholar * Schonemann, L. et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. _Genes Dev._ 28, 2381–2393 (2014). Article PubMed
PubMed Central CAS Google Scholar * Kaufmann, I., Martin, G., Friedlein, A., Langen, H. & Keller, W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates
poly(A) polymerase. _EMBO J._ 23, 616–626 (2004). Article CAS PubMed PubMed Central Google Scholar * Takagaki, Y. & Manley, J. L. RNA recognition by the human polyadenylation factor
CstF. _Mol. Cell. Biol._ 17, 3907–3914 (1997). Article CAS PubMed PubMed Central Google Scholar * Chen, F. & Wilusz, J. Auxiliary downstream elements are required for efficient
polyadenylation of mammalian pre-mRNAs. _Nucleic Acids Res._ 26, 2891–2898 (1998). Article CAS PubMed PubMed Central Google Scholar * Mandel, C. R. et al. Polyadenylation factor CPSF-73
is the pre-mRNA 3′-end-processing endonuclease. _Nature_ 444, 953–956 (2006). Article CAS PubMed Google Scholar * Bai, Y. et al. Crystal structure of murine CstF-77: dimeric association
and implications for polyadenylation of mRNA precursors. _Mol. Cell_ 25, 863–875 (2007). Article CAS PubMed Google Scholar * Yang, Q., Gilmartin, G. M. & Doublié, S. Structural
basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3′ processing. _Proc. Natl Acad. Sci. USA_ 107, 10062–10067 (2010). Article CAS PubMed
PubMed Central Google Scholar * Hunt, A. G., Xing, D. & Li, Q. Q. Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants. _BMC Genomics_ 13,
641 (2012). Article CAS PubMed PubMed Central Google Scholar * Zhang, H., Lee, J. Y. & Tian, B. Biased alternative polyadenylation in human tissues. _Genome Biol._ 6, R100 (2005).
THIS IS THE FIRST DEMONSTRATION THAT ISOFORMS USING PROXIMAL AND DISTAL PASS ARE EXPRESSED WITH BIAS IN CERTAIN TISSUES, FOR EXAMPLE IN THE BRAIN AND BLOOD. Article PubMed PubMed Central
CAS Google Scholar * Beaudoing, E. & Gautheret, D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. _Genome Res._ 11,
1520–1526 (2001). Article CAS PubMed PubMed Central Google Scholar * Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S. & Mayr, C. Ubiquitously transcribed genes use alternative
polyadenylation to achieve tissue-specific expression. _Genes Dev._ 27, 2380–2396 (2013). Article CAS PubMed PubMed Central Google Scholar * Liu, D. et al. Systematic variation in mRNA
3′-processing signals during mouse spermatogenesis. _Nucleic Acids Res._ 35, 234–246 (2007). Article CAS PubMed Google Scholar * Smibert, P. et al. Global patterns of tissue-specific
alternative polyadenylation in _Drosophila_. _Cell Rep._ 1, 277–289 (2012). Article CAS PubMed PubMed Central Google Scholar * Lee, J. Y., Ji, Z. & Tian, B. Phylogenetic analysis of
mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. _Nucleic Acids Res._ 36, 5581–5590 (2008). Article CAS PubMed PubMed Central
Google Scholar * Shepard, P. J. et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. _RNA_ 17, 761–772 (2011). Article CAS PubMed PubMed Central Google
Scholar * Dai, W. et al. A post-transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status. _Nat. Commun._ 6, 7576 (2015). Article PubMed
Google Scholar * Dass, B. et al. Loss of polyadenylation protein τCstF-64 causes spermatogenic defects and male infertility. _Proc. Natl Acad. Sci. USA_ 104, 20374–20379 (2007). Article
CAS PubMed PubMed Central Google Scholar * Sartini, B. L., Wang, H., Wang, W., Millette, C. F. & Kilpatrick, D. L. Pre-messenger RNA cleavage factor I (CFIm): potential role in
alternative polyadenylation during spermatogenesis. _Biol. Reprod._ 78, 472–482 (2008). Article CAS PubMed Google Scholar * Soumillon, M. et al. Cellular source and mechanisms of high
transcriptome complexity in the mammalian testis. _Cell Rep._ 3, 2179–2190 (2013). Article CAS PubMed Google Scholar * Zhang, P. et al. MIWI and piRNA-mediated cleavage of messenger RNAs
in mouse testes. _Cell Res._ 25, 193–207 (2015). Article CAS PubMed PubMed Central Google Scholar * Goh, W. S. et al. piRNA-directed cleavage of meiotic transcripts regulates
spermatogenesis. _Genes Dev._ 29, 1032–1044 (2015). Article CAS PubMed PubMed Central Google Scholar * Watanabe, T., Cheng, E. C., Zhong, M. & Lin, H. Retrotransposons and
pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. _Genome Res._ 25, 368–380 (2015). Article PubMed PubMed Central CAS Google Scholar * Bao, J. et al.
UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′ UTR transcripts. _PLoS Genet._ 12, e1005863 (2016). Article PubMed
PubMed Central CAS Google Scholar * Fanourgakis, G., Lesche, M., Akpinar, M., Dahl, A. & Jessberger, R. Chromatoid body protein TDRD6 supports long 3′ UTR triggered nonsense
mediated mRNA decay. _PLoS Genet._ 12, e1005857 (2016). Article PubMed PubMed Central CAS Google Scholar * Gruber, A. R. et al. Global 3′ UTR shortening has a limited effect on protein
abundance in proliferating T cells. _Nat. Commun._ 5, 5465 (2014). Article CAS PubMed Google Scholar * Fu, Y. et al. Differential genome-wide profiling of tandem 3′ UTRs among human
breast cancer and normal cells by high-throughput sequencing. _Genome Res._ 21, 741–747 (2011). Article CAS PubMed PubMed Central Google Scholar * Morris, A. R. et al. Alternative
cleavage and polyadenylation during colorectal cancer development. _Clin. Cancer Res._ 18, 5256–5266 (2012). Article CAS PubMed Google Scholar * Flavell, S. W. et al. Genome-wide
analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. _Neuron_ 60, 1022–1038 (2008). Article CAS PubMed
PubMed Central Google Scholar * Chang, J. W. et al. mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. _Nat. Commun._ 6, 7218 (2015). Article CAS PubMed Google
Scholar Download references ACKNOWLEDGEMENTS The authors thank members of their laboratories for helpful discussions, and I. Boluck for assistance with manuscript preparation. Work in the
authors' laboratories was funded by grants GM84089 (B.T.), and GM28983 and GM118136 (J.L.M.) from the US National Institutes of Health. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS *
Department of Microbiology, Biochemistry and Molecular Genetics, New Jersey Medical School, Rutgers University, Newark, 07103, New Jersey, USA Bin Tian * Department of Biological Sciences,
Columbia University, New York, 10027, New York, USA James L. Manley Authors * Bin Tian View author publications You can also search for this author inPubMed Google Scholar * James L. Manley
View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Bin Tian or James L. Manley. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION S1 (BOX) Variation in alternative polyadenylation across species (PDF 122
kb) POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 GLOSSARY * PUF protein (Pumilio and FBF homology family protein). A member of a
family of RNA-binding proteins that regulate aspects of mRNA metabolism by binding to specific sequences in 3′ untranslated regions. * STAU1-mediated mRNA decay An mRNA decay mechanism in
which RNA structures in the 3′ untranslated region interact with double-stranded RNA-binding protein Staufen homologue 1 (STAU1) to mediate mRNA decay. * AU-rich element-mediated decay mRNA
decay elicited by the presence of AU-rich elements (AREs) in the 3′ untranslated region. * PIWI-interacting RNAs Small non-coding RNAs that form RNA–protein complexes with PIWI proteins to
silence transposable elements in germline cells of metazoans. * Non-stop decay An mRNA decay mechanism that specifically degrades mRNAs without a stop codon. * Exosome A nuclear or
cytoplasmic multiprotein complex that degrades mRNAs through the activity of 3′-to-5′ exoribonucleases. * Non-canonical PAPs (Non-canonical poly(A) polymerases). Enzymes that have distinct
structural features and are capable of synthesizing poly(A) tails but are not typically associated with the polyadenylation machinery. * Paused Pol II (Paused RNA polymerase II). Pol II that
has paused in the promoter-proximal region of the mRNA and is poised for productive elongation. * Paraspeckle A dynamic nuclear compartment composed of RNA-binding proteins and RNAs. The
functions of paraspeckles are not entirely clear. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Tian, B., Manley, J. Alternative polyadenylation of
mRNA precursors. _Nat Rev Mol Cell Biol_ 18, 18–30 (2017). https://doi.org/10.1038/nrm.2016.116 Download citation * Published: 28 September 2016 * Issue Date: January 2017 * DOI:
https://doi.org/10.1038/nrm.2016.116 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative