- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
KEY POINTS * The tagging of proteins with ubiquitin and their subsequent degradation by the 26S proteasome provides a highly regulated proteolysis system that is necessary in all eukaryotes.
* The ubiquitylation of a target protein involves the sequential action of E1, E2 and E3 enzymes; E3 ubiquitin ligase provides target specificity. * Plants use ubiquitin and the 26S
proteasome to control a wide variety of developmental processes through the degradation of key cellular regulators. * Approximately 5% of the _Arabidopsis thaliana_ genome encodes components
of the ubiquitin/26S proteasome system. * We discuss the roles of ubiquitin in the regulation of floral development, responses to plant hormones and to pathogens, and in the regulation of
photomorphogenesis. * The COP9 signalosome (CSN) is intimately connected to the ubiquitin/26S proteasome system and might regulate the activity of SCF-type (and possibly other) E3 ligases. *
Recent advances, particularly in the fields of hormone signalling and responses to pathogens, have shown just how frequently plants use ubiquitin to regulate cellular processes and indicate
that ubiquitin and the 26S proteasome act as regulators in many more, as yet unidentified, plant developmental processes. ABSTRACT A tightly regulated and highly specific system for the
degradation of individual proteins is essential for the survival of all organisms. In eukaryotes, this is achieved by the tagging of proteins with ubiquitin and their subsequent recognition
and degradation by the 26S proteasome. In plants, genetic analysis has identified many genes that regulate developmental pathways. Subsequent analysis of these genes has implicated ubiquitin
and the 26S proteasome in the control of diverse developmental processes, and indicates that proteolysis is a crucial regulatory step throughout the life cycle of plants. Access through
your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12
print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be
subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR
CONTENT BEING VIEWED BY OTHERS THE RPN12A PROTEASOME SUBUNIT IS ESSENTIAL FOR THE MULTIPLE HORMONAL HOMEOSTASIS CONTROLLING THE PROGRESSION OF LEAF SENESCENCE Article Open access 30
September 2022 CUL3 E3 LIGASES IN PLANT DEVELOPMENT AND ENVIRONMENTAL RESPONSE Article 15 January 2021 MECHANISMS CONTROLLING PLANT PROTEASES AND THEIR SUBSTRATES Article 08 February 2023
REFERENCES * Tanaka, K. Molecular biology of proteasomes. _Mol. Biol. Rep._ 21, 21–26 (1995). CAS PubMed Google Scholar * Hochstrasser, M. Ubiquitin, proteasomes, and the regulation of
intracellular protein degradtion. _Curr. Biol._ 7, 215–223 (1995). CAS Google Scholar * Vierstra, R. D. Proteolysis in plants: mechanisms and functions. _Plant Mol. Biol._ 32, 275–302
(1996). CAS PubMed Google Scholar * Vierstra, R. D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. _Trends Plant Sci._ 8, 135–142
(2003). CAS PubMed Google Scholar * Hellmann, H. & Estelle, M. Plant development: regulation by protein degradation. _Science_ 297, 793–797 (2002). CAS PubMed Google Scholar *
Frugis, G. & Chua, N. H. Ubiquitin-mediated proteolysis in plant hormone signal transduction. _Trends Cell Biol._ 12, 308–311 (2002). CAS PubMed Google Scholar * Pickart, C. M.
Mechansims underlysing ubiquitination. _Ann. Rev. Biochem._ 70, 503–533 (2001). CAS PubMed Google Scholar * Bachmir, A., Novatchkova, M., Potushak, T. & Eisenhaber, F. Ubiquitylation
in plants: a post-genic look at post-translational modification. _Trends Plant Sci._ 6, 463–470 (2001). Google Scholar * Aguilar, R. C. & Wendland, B. Ubiqutin: not just for proteasomes
anymore. _Curr. Opin. Cell Biol._ 15, 184–190 (2003). CAS PubMed Google Scholar * Conaway, R. C., Brower, C. S. & Weliky-Conaway, J. Emerging roles of ubiquitin in transcriptional
regulation. _Science_ 296, 1254–1258 (2002). CAS PubMed Google Scholar * Weissmann, A. M. Themes and variations on ubiquitylation. _Nature Rev. Mol. Cell Biol._ 2, 169–178 (2001). Google
Scholar * Serino, G. & Deng, X. W. The COP9 signalosome: regulating plant development through the control of proteolysis. _Ann. Rev. Plant Biol._ 54, 165–182 (2003). CAS Google Scholar
* Bech-Otschir, D., Seeger, M. & Dubiel, W. The COP9 signalosome: at the interface between signal transduction and ubiquitin dependent proteolysis. _J. Cell Sci._ 115, 467–473 (2002).
CAS PubMed Google Scholar * Schwechheimer, C. & Deng, X. W. COP9 signalosome revisited; a novel mediator of protein degradation. _Trends Cell Biol._ 11, 420–426 (2001). CAS PubMed
Google Scholar * Peng, Z. et al. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases _in vivo_. _Curr. Biol._ 13, R504–R505 (2003). CAS
PubMed Google Scholar * Lyapina, S. et al. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. _Science_ 292, 1382–1385 (2001). CAS PubMed Google Scholar * Schwechheimer,
C. et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. _Science_ 292, 1379–1382 (2001). SHOWS THAT REDUCED CSN FUNCTION LEADS TO A
LOSS OF AUXIN RESPONSES SIMILAR TO THAT SEEN IN MUTANTS IN SCFTIR1. THEY ALSO SHOW THAT THE CSN SCFTIR1 INTERACT _IN VIVO_ AND THAT THE CSN IS REQUIRED FOR THE DEGRADATION OF A PUTATIVE
TARGET OF SCFTIR1, INDICATING THE IMPORTANCE OF THE CSN IN REGULATING SCF-TYPE E3 LIGASES. CAS PubMed Google Scholar * Cope, G. A. et al. Role of predicted metalloprotease motif of
JAB1/Csn5 in cleavage of Nedd8 from Cul1. _Science_ 298, 608–611 (2002). THE FIRST DEMONSTRATION THAT THE JAB1/CSN5 SUBUNIT OF THE CSN SHOWS NEDD8 ISOPEPTIDASE ACTIVITY ASSOCIATED WITH ITS
METALLOENZYME JAMM DOMAIN. CAS PubMed Google Scholar * Leyser, H. M. O. et al. _Arabidopsis_ auxin-resistance gene _AXR1_ encodes a protein related to ubiquitin activating enzyme E1.
_Nature_ 364, 161–164 (1993). CAS PubMed Google Scholar * del Pozo, J. C., Timpte, C., Tan, S., Callis, J. & Estelle, M. The ubiquitin-related protein RUB1 and auxin response in
_Arabidopsis_. _Science_ 280, 1760–1763 (1998). CAS Google Scholar * Schwechheimer, C., Serino, G. & Deng, X. W. Multiple ubiquitin ligase-mediated processes require COP9 signalosome
and AXR1 function. _Plant Cell_ 14, 2553–2563 (2002). SHOWS THE ROLE OF THE CSN IN REGULATING MANY SCF E3 LIGASES IN PLANTS. CAS PubMed PubMed Central Google Scholar * Estelle, M.
Proteases and cellular regulation in plants. _Curr. Opin. Plant Biol._ 4, 254–260 (2001). CAS PubMed Google Scholar * Adam, Z. Chloroplast proteases; possible regulators of gene
expression? _Biochimie_ 82, 647–654 (2000). CAS PubMed Google Scholar * Palma, J. M. et al. Plant proteases, protein degradtion, and oxidative stress; role of peroxisomes. _Plant Physiol.
Biochem._ 40, 521–530 (2002). CAS Google Scholar * Adam, Z. & Clarke, A. K. Cutting edge of chloroplast proteolysis. _Trends Plant Sci._ 7, 451–456 (2002). CAS PubMed Google Scholar
* Woo, R. H. et al. ORE9, an F-box protein that regulates leaf senescence in _Arabidopsis_. _Plant Cell_ 13, 1779–1790 (2001). CAS PubMed PubMed Central Google Scholar * Stirnberg, P.,
van de Sande, K. & Leyser, H. M. O. _MAX_ and _MAX2_ control shoot lateral branching in _Arabidopsis_. _Development_ 129, 1131–1141 (2002). CAS PubMed Google Scholar * Schultz, T.
F., Kiyosue, T., Yanovsky, M., Wada, M. & Kay, S. A. A role for LKP2 in the circadian clock of _Arabidopsis_. _Plant Cell_ 13, 2659–2670 (2001). CAS PubMed PubMed Central Google
Scholar * Millar, A. J., Carré, I. A., Strayer, C. A., Chua, N. -H. & Kay, S. A. Circadian clock mutants in _Arabidopsis_ identified by luciferase imaging. _Science_ 267, 1161–1163
(1995). CAS PubMed Google Scholar * Kim, A. S. & Delaney, T. P. _Arabidopsis_ SON1 is an F-box protein that regulates a novel induced defense response independent of both salicyclic
acid and systemic acquired resistance. _Plant Cell_ 14, 1469–1482 (2002). CAS PubMed PubMed Central Google Scholar * Thelander, M., Fredriksson, D., Schouten, J., Hoge, H. C. &
Ronne, H. Cloning by pathway activation in yeast: identification of an _Arabidopsis thaliana_ F-box protein that can turn on glucose repression. _Plant Mol. Biol._ 49, 69–79 (2002). CAS
PubMed Google Scholar * Ingram, G. C. et al. Dual role for _fimbriata_ in regulating floral homeotic genes and cell division in _Antirrhinum_. _EMBO J._ 16, 6521–6534 (1997). CAS PubMed
PubMed Central Google Scholar * Karlowski, W. M. & Hirsch, A. M. The overexpression of an alfalfa RING-H2 gene induces pleiotrpic effects on plant growth and development. _Plant Mol.
Biol._ 52, 121–133 (2003). CAS PubMed Google Scholar * Stone, S. L., Anderson, E. M., Mullen, R. T. & Goring, D. R. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of
proteins during the rejection of self-incompatible _Brassica_ pollen. _Plant Cell_ 15, 885–898 (2003). DEMONSTRATES THAT, IN THE PRESENCE OF INCOMPATIBLE POLLEN, THE E3 LIGASE ARC1 SHUTTLES
BETWEEN THE NUCLEUS AND THE CYTOSOL AND PROMOTES THE PROTEIN DEGRADATION EVENTS THAT LEAD TO POLLEN REJECTION. PubMed PubMed Central Google Scholar * Downes, B. P., Stupar, R. M.,
Gingerich, D. J. & Vierstra, R. D. The HECT ubiquitin-protein ligase (UPL) family in _Arabidopsis_; UPL3 has a specific role in trichome development. _Plant J._ 35, 729–742 (2003).
DESCRIBES ANALYSIS OF KNOCKOUT LINES FOR THE _ARABIDOPSIS_ HECT-DOMAIN PROTEINS UPL3 AND UPL4. SHOWS THAT MUTATIONS IN _UPL3_ LEAD TO DEFECTS IN TRICHOME DEVELOPMENT AND THAT _UPL3_ IS
ALLELIC TO THE PREVIOUSLY IDENTIFIED _KAKTUS_ MUTANT. CAS PubMed Google Scholar * Irish, V. F. Patterning the flower. _Dev. Biol._ 209, 211–220 (1999). CAS PubMed Google Scholar *
Yanofsky, M. F. Floral meristems to floral organs; genes controlling early events in _Arabidopsis_ flower development. _Ann. Rev. Plant Physiol. Plant Mol. Biol._ 46, 167–188 (1995). CAS
Google Scholar * Coen, E. S. & Meyerowitz, E. M. The war of the whorls; genetic interactions controlling flower development. _Nature_ 353, 31–37 (1991). CAS PubMed Google Scholar *
Haughn, G. W. & Somerville, C. R. Genetic control of morphogenesis in _Arabidopsis_. _Dev. Genet._ 9, 73–89 (1988). Google Scholar * Riechmann, J. L. & Meyerowitz, E. MADS domain
proteins in plant development. _J. Biol. Chem._ 378, 1079–1101 (1997). CAS Google Scholar * Goto, K. & Meyerowitz, E. M. Function and regulation of the _Arabidopsis_ floral homeotic
gene _PISTILLATA_. _Genes Dev._ 8, 1548–1560 (1994). CAS PubMed Google Scholar * Jack, T., Brockman, L. L. & Meyerowitz, E. M. The homeotic gene _APETALA3_ of _Arabidopsis thaliana_
encodes a MADS box and is expressed in petals and stamens. _Cell_ 68, 683–697 (1992). CAS PubMed Google Scholar * Jack, T., Fox, G. L. & Meyerowitz, E. M. _Arabidopsis_ homeotic gene
_APETALA3_ ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. _Cell_ 76, 703–716 (1994). CAS PubMed Google Scholar * Krizek, B. A.
& Meyerowitz, E. M. The _Arabidopsis_ homeotic genes _APETALA3_ and _PISTILLATA_ are sufficient to provide the B class organ identity function. _Development_ 122, 11–22 (1996). CAS
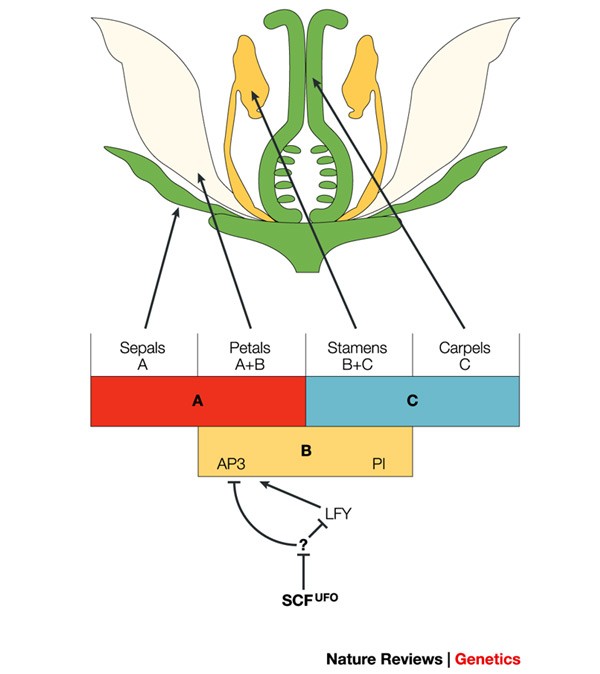
PubMed Google Scholar * Levin, J. Z. & Meyerowitz, E. M. _UFO:_ an _Arabidopsis_ gene involved in both floral meristem and floral organ development. _Plant Cell_ 7, 529–548 (1995). CAS
PubMed PubMed Central Google Scholar * Wilkinson, M. D. & Haughn, G. W. _UNUSUAL FLORAL ORGANS_ controls meristem identity and organ primordial fate in _Arabidopsis_. _Plant Cell_
7, 1485–1499 (1995). CAS PubMed PubMed Central Google Scholar * Lee, I., Wolfe, D. S., Nilsson, O. & Weigel, D. A. _LEAFY_ co-regulator encoded by _UNUSUAL FLORAL ORGANS_. _Curr.
Biol._ 7, 95–104 (1997). PubMed Google Scholar * Zhao, D., Yu, Q., Chen, M. & Ma, H. The _ASK1_ gene regulates B function gene expression in cooperation with _UFO_ and _LEAFY_ in
_Arabidopsis_. _Development_, 128, 2735–2746 (2001). CAS PubMed Google Scholar * Samach, A. et al. The _UNUSUAL FLORAL ORGANS_ gene of _Arabidopsis thaliana_ is an F-box protein required
for normal patterning and growth in the floral meristem. _Plant J._ 20, 433–445 (1999). CAS PubMed Google Scholar * Wang, X. et al. The COP9 signalosome interacts with SCFUFO and
participates in _Arabidopsis_ flower development. _Plant Cell_ 15, 1071–1082 (2003). SHOWS THAT THE CSN IS HIGHLY ENRICHED IN FLOWERS AND THAT PLANTS WITH REDUCED CSN FUNCTION SHOW DEFECTS
IN FLORAL DEVELOPMENT. IT ALSO SHOWS THAT THE F-BOX PROTEIN, UFO, FORMS PART OF AN SCF-TYPE E3 LIGASE THAT ASSOCIATES WITH THE CSN _IN VIVO_. CAS PubMed PubMed Central Google Scholar *
Peng, Z., Serino, G. & Deng, X. W. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in _Arabidopsis_. _Plant Cell_ 13,
2393–2407 (2001). CAS PubMed PubMed Central Google Scholar * Johri, M. M. & Mitra, D. Action of plant hormones. _Curr. Sci._ 80, 199–205 (2001). CAS Google Scholar * Vogler, H.
& Kuhlemeier, C. Simple hormones but complex signaling. _Curr. Opin. Plant Biol._ 6, 51–56 (2003). CAS PubMed Google Scholar * Leyser, O. Molecular genetics of auxin signaling. _Ann.
Rev. Plant Biol._ 53, 377–398 (2002). CAS Google Scholar * Rogg, L. E. & Bartel, B. Auxin signaling: derepression through regulated proteolysis. _Dev. Cell_ 1, 595–604 (2001). CAS
PubMed Google Scholar * del Pozo, J. C. & Estelle, M. Function of the ubiquitin-proteasome pathway in auxin response. _Trends Plant Sci._ 4, 107–112 (1999). PubMed Google Scholar *
Richards, D. E., King, K. E., Ait-ali, T. & Harberd, N. P. How gibberellin regulates plant growth and development: a molecular genetic analysis of giberellin signaling. _Ann. Rev. Plant
Physiol. Plant Mol. Biol._ 52, 67–88 (2001). CAS Google Scholar * Koornneef, M. & van der Veen, J. H. Induction and analysisof gibberellin sensitive mutants in _Arabidopsis thaliana_
(L.) Heynh. _Theor. Appl. Genet._ 58, 257–263 (1980). CAS PubMed Google Scholar * Sun, T., Goodman H. M. & Ausubel, F. M. Cloning the _Arabidopsis GA1_ locus by genomic subtraction.
_Plant Cell_ 4, 119–128 (1992). CAS PubMed PubMed Central Google Scholar * Fath, A., Bethke, P., Lonsdale, J., Meza-Romero, R. & Jones, R. Programmed cell death in cereal aleurone.
_Plant Mol. Biol._ 44, 255–266 (2000). CAS PubMed Google Scholar * Silverstone, A. L., Ciampaglio, C. N. & Sun, T. The _Arabidopsis RGA_ gene encodes a transcriptional regulator
repressing the gibberellin signal transduction pathway. _Plant Cell_ 10, 155–169 (1998). CAS PubMed PubMed Central Google Scholar * Silverstone, A. L. et al. Repressing a repressor:
gibberellin-induced rapid reduction of the RGA protein in _Arabidopsis_. _Plant Cell_ 13, 1555–1565 (2001). CAS PubMed PubMed Central Google Scholar * Itoh, H., Ueguchi-Tanaka, M., Sato,
Y., Ashikari, M. & Matsuoka, M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. _Plant Cell_ 14, 57–70 (2002). CAS PubMed
PubMed Central Google Scholar * Fu, X. et al. Gibberellin-mediated proteasome-dependent degradtion of the barley DELLA protein SLN1 repressor. _Plant Cell_ 14, 3191–3200 (2002). CAS
PubMed PubMed Central Google Scholar * Sasaki, A. et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. _Science_ 299, 1896–1989 (2003).
DEMONSTRATES THAT THE GA-INSENSITIVE MUTANT _GID2_ OF RICE IS CAUSED BY A MUTATION IN AN F-BOX PROTEIN THAT FORMS PART OF AN SCF-TYPE E3 LIGASE. THEY ALSO SHOW THAT DEGRADATION OF SLR1, A
REPRESSOR OF GA SIGNALLING, INVOLVES SCFGID2 AND IS CONTROLLED THROUGH GA-DEPENDENT PHOSOPHORYLATION OF SLR1. CAS PubMed Google Scholar * McGinnis, K. M. et al. The _Arabidopsis SLEEPY1_
gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. _Plant Cell_ 15, 1120–1130 (2003). IDENTIFIES THE _ARABIDOPSIS SLEEPY_ GENE, A POSITIVE REGULATOR OF GA RESPONSES. SLEEPY
IS AN F-BOX PROTEIN THAT FORMS AN SCF E3 LIGASE AND PARTICIPATES IN THE DEGRADATION OF THE REPRESSOR OF GA SIGNALLING RGA IN RESPONSE TO GA. CAS PubMed PubMed Central Google Scholar *
Wang, K. L., Li, H. & Ecker, J. R. Ethylene biosynthesis and siganling networks. _Plant Cell Suppl._ 14, S131–S151 (2002). CAS Google Scholar * Yanagisawa, S., Yoo, S. D. & Sheen,
J. Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. _Nature_ 425, 521–525 (2003). DEMONSTRATES THAT EIN3, A CRUCIAL REGULATOR OF ETHYLENE SIGNALLING, IS
STABILIZED BY ETHYLENE. BY CONTRAST, ADDITION OF GLUCOSE CAUSES THE DEGRADATION OF EIN3 IN A PROTEASOME-DEPENDENT MANNER, INDICATING THAT EIN3 IS A CONVERGENCE POINT BETWEEN ETHYLENE AND
SUGAR SIGNALLING. CAS PubMed Google Scholar * Wang, Z. Y. et al. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. _Nature_ 410, 380–383 (2001). CAS PubMed
Google Scholar * Li, J. & Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. _Science_ 288, 2360–2363 (1997). Google Scholar *
Wang, Z. Y. et al. Nuclear-localised BZR1 mediates brassionosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. _Dev. Cell_ 2, 505–513 (2002). CAS PubMed
Google Scholar * He, J. -X., Gendron, J. M., Yang, Y. & Wang, Z. -Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid
signaling pathway in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 99, 10185–10190 (2002). CAS PubMed PubMed Central Google Scholar * Lopez-Molina, L., Mongrand, S. & Chua, N. H. A
post-germination developmental arrest checkpont is mediated by abscisic acid and requires the ABI5 transcription factor in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 32, 1–12 (2001).
DESCRIBES THE IDENTIFICATION OF ABI5, A TRANSCRIPTION FACTOR THAT MEDIATES RESPONSES TO ABA. THEY ALSO SHOW THAT ABI5 IS REGULATED AT THE LEVEL OF PROTEIN DEGRADATION AND REQUIRES THE ACTION
OF THE 26S PROTEASOME AND THE ABI5 BINDING PROTEIN, AFP. Google Scholar * Lopez-Molina, L., Mongrand, S., Kinoshita, N. & Chua, N. H. AFP is a novel negative regulator of ABA signaling
that promotes ABI5 protein degradation. _Genes Dev._ 17, 410–418 (2003). CAS PubMed PubMed Central Google Scholar * Zhang, S. & Klessig, D. F. MAPK cascades in plant defense
signaling. _Trends Plant Sci._ 6, 520–527 (2001). CAS PubMed Google Scholar * Dong, X. N. Genetic dissection of systemic acquired resistance. _Curr. Opin. Plant Biol._ 4, 309–314 (2001).
CAS PubMed Google Scholar * McDowell, J. M. & Dangl, J. L. Signal transduction in the plant immune response. _Trends Biochem. Sci._ 25, 79–82 (2000). CAS PubMed Google Scholar *
Shirasu, K. & Schulze-Lefert, P. Regulators of cell death in disease resistance. _Plant Mol. Biol._ 44, 371–385 (2000). CAS PubMed Google Scholar * Dong, X. & Sa, J. A. Ethylene
and disease resistance in plants. _Curr. Opin. Plant Biol._ 1, 316–323 (1988). Google Scholar * Hammond-Kosack, K. E. & Parker, J. E. Deciphering plant-pathogen communication: fresh
perspectives for molecular resistance breeding. _Curr. Opin. Biotechnol._ 14, 177–193 (2003). CAS PubMed Google Scholar * Dangl, J. L. & Jones, J. D. G. Plant pathogens and integrated
defense responses to infection. _Nature_ 411, 826–833 (2001). CAS PubMed Google Scholar * Shirasu, K. et al. A novel class of eukaryotic zinc-finger binding protein is required for
disease resistance signaling in barley and development in _C. elegans_. _Cell_ 99, 355–366 (1999). CAS PubMed Google Scholar * Azevedo, C. et al. The RAR1 interactor SGT1, an essential
component of R gene-triggered disease resistance. _Science_ 295, 2073–2076 (2002). SHOWS THAT SGT1 IS AN INTERACTING PARTNER OF RAR1, AN ESSENTIAL COMPONENT OF MLA12-MEDIATED RESISTANCE
AGAINST POWDERY MILDEW IN BARLEY. SINGLE-CELL RNAI ASSAYS SHOWED THAT SGT1 IS ALSO A CRUCIAL COMPONENT OF THE RESISTANCE PATHWAY. CAS PubMed Google Scholar * Austin, M. J. et al.
Regulatory role of _SGT1_ in early R gene-mediated plant defenses. _Science_ 295, 2077–2080 (2002). DESCRIBES A FORWARD MUTANT SCREENING IN _ARABIDOPSIS_ FOR LOSS OF RESISTANCE AGAINST DOWNY
MILDEW THAT IDENTIFIED SGT1B AS A VITAL COMPONENT OF RESISTANCE. CAS PubMed Google Scholar * Peart, J. R. et al. Ubiquitin ligase associated protein SGT1 is required for host and
non-host disease resistance in plants. _Proc. Natl Acad. Sci. USA_ 99, 10865–10869 (2002). CAS PubMed PubMed Central Google Scholar * Shirasu, K. & Schulze-Lefeft, P. Complex
formation, promiscuity and multi-functionality: protein interactions in disease resistance pathways. _Trends Plant Sci._ 8, 252–258 (2003). CAS PubMed Google Scholar * Takahashi, A.,
Casais, C., Ichimura, K. & Shirasu, K. HSP90 interacts with RAR1 and SCT1, and is essential for RPS2-mediated disease resistance in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 100,
11777–11782 (2003). CAS PubMed PubMed Central Google Scholar * Dubacq, C., Guerois, R., Courbeyrette, R., Kitagawa, K. & Mann, C. Sgt1p contributes to cyclic AMP pathway activity and
physically interacts with the adenylyl cyclase Cry1p/Cdc35p in budding yeast. _Eukaryot. Cell_ 1, 568–582 (2002). CAS PubMed PubMed Central Google Scholar * Liu, Y., Schiff, M., Serino,
G., Deng, X. W. & Dinesh-Kumar, S. P. Role of SCF ubiquitin-ligase and the COP9 signalosome in the _N_ gene-mediated resistance response to _Tobacco mosaic virus_. _Plant Cell_ 14,
1483–1496 (2002). SHOWS THAT SKP1 AND CSN COMPONENTS ARE ESSENTIAL FOR N-MEDIATED DISEASE RESISTANCE IN _NICOTIANA_ . BOTH RAR1 AND SGT1 ASSOCIATE WITH SCF AND CSN COMPONENTS, INDICATING THE
FUNCTIONAL LINK AMONG THESE COMPONENTS. CAS PubMed PubMed Central Google Scholar * Kitagawa, K., Skowyra, D., Elledge, S. J., Harper, J. W. & Hieter, P. SGT1 encodes an essential
component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. _Mol. Cell_ 4, 21–33 (1999). CAS PubMed Google Scholar * Gray, W. M., Muskett,
P. R., Chuang, H. -W. & Parker, J. E. _Arabidopsis_ SGT1b is required for SCFTIR1-mediated auxin response. _Plant Cell_ 15, 1310–1319 (2003). CAS PubMed PubMed Central Google Scholar
* Creelmann, R. A. & Mullet, J. E. Biosynthesis and action of jasmonates in plants. _Ann. Rev. Plant Physiol. Plant Mol. Biol._ 48, 355–381 (1997). Google Scholar * Feys, B. J. F.,
Benedetti, C. E., Penfold, C. N. & Turner, J. G. _Arabidopsis_ mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and
resistant to a bacterial pathogen. _Plant Cell_ 6, 751–759 (1994). CAS PubMed PubMed Central Google Scholar * Thomma, B. P. H. J. et al. Separate jasmonate-dependent and
salicylate-dependent defense-repsonse pathways in _Arabidopsis_ are essential for resistance to distinct microbial pathogens. _Proc. Natl Acad. Sci. USA_ 95, 15107–15111 (1998). CAS PubMed
PubMed Central Google Scholar * McConn, M., Creelmann, R. A., Bell, E., Mullet, J. E. & Browse, J. Jasmonate is essential for insect defense. _Proc. Natl Acad. Sci. USA_ 94,
5473–5477 (1997). CAS PubMed PubMed Central Google Scholar * Devoto, A. et al. COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in _Arabidopsis_. _Plant
J._ 32, 457–466 (2002) CAS PubMed Google Scholar * Xu, L. et al. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in _Arabidopsis_. _Plant Cell_ 14, 1919–1935
(2002). CAS PubMed PubMed Central Google Scholar * Xie, D., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. _COI1_: an _Arabidopsis_ gene required for jasmonate-regulated
defense and fertility. _Science_ 280, 1091–1094 (1998). A LANDMARK PAPER DESCRIBING ISOLATION OF THE _COI1_ GENE THAT ENCODES AN F-BOX PROTEIN, REQUIRED FOR JASMONATE, THE SIGNALLING PATHWAY
IN _ARABIDOPSIS_. CAS PubMed Google Scholar * Feng, S. et al. The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. _Plant Cell_ 15, 1083–1094 (2003).
CAS PubMed PubMed Central Google Scholar * Sullivan, J. A. & Deng, X. W. From seed to seed; the role of photoreceptors in _Arabidopsis_ development. _Dev. Biol._ 260, 289–297
(2003). CAS PubMed Google Scholar * Quail, P. H. Phytochrome photosensory signaling networks. _Nature Rev. Mol. Cell Biol._ 3, 85–93 (2002). CAS Google Scholar * Wang, H. & Deng, X.
W. in _The Arabidopsis Book_, American Society of Plant Biologists [online], (eds Somerville, C. R. & Meyerowitz, E. M.), (cited 29 Oct 2003)
<http://www.aspb.org/publications/arabidopsis> (2002). Google Scholar * Teppermann, J. M., Zhu, T., Chang, H. -S., Wang, X. & Quail, P. H. Multiple transcription factor
genes are early targets of phytochrome A signaling. _Proc. Natl Acad. Sci. USA_ 98, 9437–9442 (2001). Google Scholar * Ma, L. et al. Light control of _Arabidopsis_ development entails
coordinated regulation of genome expression and cellular pathways. _Plant Cell_ 13, 2589–2398 (2001). CAS PubMed PubMed Central Google Scholar * Lin, C. Blue light photoreceptors and
signal transduction. _Plant Cell_ 14, S207–S225 (2002). CAS PubMed PubMed Central Google Scholar * Möller, S. G., Ingles, P. J. & Whitelam, G. C. The cell biology of phytochrome
signaling. _New Phytol._ 154, 553–590 (2002). PubMed Google Scholar * Wei, N. & Deng, X. W. The role of the _COP/DET/FUS_ genes in light control of _Arabidopis_ seedling development.
_Plant Physiol._ 112, 871–878 (1996). CAS PubMed PubMed Central Google Scholar * Deng, X. W. et al. COP1, an _Arabidopsis_ regulatory gene, encodes a protein with both a zinc-binding
motif and a Gb homologous domain. _Cell_ 71, 791–801 (1992). CAS PubMed Google Scholar * Pepper, A., Delaney, T., Washburn, T., Poole, D. & Chory, J. _DET1_, a negative regulator of
light-mediated development and gene expression in _Arabidopsis_, encodes a novel nuclear localized protein. _Cell_ 78, 109–116 (1994). CAS PubMed Google Scholar * Suzuki, G., Yanagawa,
Y., Kwok, S. F. & Deng, X. W. _Arabidopsis_ COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. _Genes
Dev._ 16, 554–559 (2002). DESCRIBES THE IDENTIFICATION OF _COP10_ , ONE OF THE ESSENTIAL COP/DET/FUS GENES. _COP10_ ENCODES A PROTEIN SHOWING SIGNIFICANT HOMOLOGY TO E2 VARIANT PROTEINS AND
INDICATES A POSSIBLE ROLE FOR NON-CANONICAL UBIQUITYLATION IN THE REGULATION OF PHOTOMORPHOGENESIS. CAS PubMed PubMed Central Google Scholar * Wei, N., Chamovitz, D. A. & Deng, X.
W. _Arabidopsis_ COP9 is a component of a novel signaling complex mediating light control of development. _Cell_ 78, 117–124 (1994). CAS PubMed Google Scholar * Holm, M., Ma, L. -G., Qu,
L. -J. & Deng, X. W. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in _Arabidopsis_. _Gene Dev._ 16, 1247–1259 (2002). CAS
PubMed PubMed Central Google Scholar * Ballesteros, M. et al. LAF1, a myb transcription activator for phytochrome A signaling. _Genes Dev._ 15, 2613–2625 (2001). CAS PubMed PubMed
Central Google Scholar * Ang, L. -H. et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of _Arabidopsis_ development. _Mol. Cell_ 1, 213–222
(1998). CAS PubMed Google Scholar * Oyama, T., Shimura, Y. & Okada, K. The _Arabidopsis HY5_ gene encodes a bZIP protein that regulates stimulus-induced development of root and
hypocotyl. _Genes Dev._ 11, 2983–2995 (1997). CAS PubMed PubMed Central Google Scholar * von Arnim, A. G. & Deng, X. W. Light inactivation of _Arabidopsis_ photomorphogenic repressor
COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. _Cell_ 79, 1035–1045 (1994). CAS PubMed Google Scholar * Osterlund, M. T., Hardtke, C., Wei, N. &
Deng, X. W. Targeted destabilization of Hy5 during light-regulated development of _Arabidopsis_. _Nature_ 405, 462–466 (2000). A LANDMARK PAPER THAT SHOWS THAT THE PHOTOMORPHOGENESIS
PROMOTING FACTOR HY5 IS REGULATED AT THE LEVEL OF PROTEIN DEGRADATION THROUGH THE 26S PROTEASOME AND THAT THIS REGULATION REQUIRES THE ACTION OF COP1 AND OTHER COP/DET/FUS PROTEINS. CAS
PubMed Google Scholar * Saijo, Y. et al. The COP1–SPA1 interaction defines a critical step in Phytochrome A-mediated regulation of HY5 activity. _Genes Dev._ 17, 2642–2647 (2003). SHOWS
THAT COP1 HAS E3-LIGASE ACTIVITY WITH RESPECT TO HY5 AND THAT THIS ACTIVITY CAN BE ALTERED BY SPA1, AN UPSTREAM REGULATOR OF PHYTOCHROME A SIGNALLING. CAS PubMed PubMed Central Google
Scholar * Seo, S. H. et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. _Nature_ 423, 995–999 (2003). THIS IS THE FIRST DEMONSTRATION OF THE E3
ACTIVITY OF COP1 ON THE TRANSCRIPTION FACTOR, LAF1, AND SHOWS THAT THIS ACTIVITY CAN BE STIMULATED BY ADDITION OF THE COP1-BINDING REGION OF SPA1. CAS PubMed Google Scholar * Hoffmann, R.
M. & Pickart, C. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. _Cell_ 96, 645–653 (1999). Google Scholar *
Benvenuto, G., Formiggini, F., Laflamme, P., Malakhov, M. & Bowler, C. The photomorphogensis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosomal context. _Curr.
Biol._ 12, 1529–1534 (2002). CAS PubMed Google Scholar * Schroeder, D. F. et al. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate _Arabidopsis_ phoromorphogenesis.
_Curr. Biol._ 15, 1462–1472 (2002). Google Scholar * Chamovitz, D. A. et al. The COP9 complex a novel multisubunit nuclear regulator involved in light control of a plant developmental
switch. _Cell_ 86, 115–121 (1996). CAS PubMed Google Scholar * Quail, P. An emerging molecular map of the phytochromes. _Plant Cell Environ._ 20, 657–666 (1997). CAS Google Scholar *
Wang, H., Ma, L., Li, J., Zhao, H. & Deng, X. W. Direct interaction of _Arabidopsis_ cryptochromes with COP1 in light control development. _Science_ 294, 154–158 (2001). CAS PubMed
Google Scholar * Yang, H. -Q., Tang, R. -H. & Cashmore, A. R. The signaling mechanism if _Arabidopsis_ CRY1 involves direct interaction with COP1. _Plant Cell_ 13, 2573–2587 (2001). CAS
PubMed PubMed Central Google Scholar * Marx, J. Ubiquitin lives up to its name. _Science_ 297, 1792–1794 (2002). PubMed Google Scholar * Staub, J. M., Wei, N. & Wang, X. W.
Evidence for FUS6 as a component of the nuclear localized COP9 complex in _Arabidopsis_. _Plant Cell_ 8, 2047–2056 (1996). CAS PubMed PubMed Central Google Scholar * Serino, G. et al.
Characterization of the last subunit of the _Arabidopsis_ COP9 signalosome: implications for the overall structure and origian of the complex. _Plant Cell_ 15, 719–731 (2003). CAS PubMed
PubMed Central Google Scholar * Peng, Z., Serino, G. & Deng, X. W. A role of _Arabidopsis_ COP9 signalosome in multifaceted developmental processes revealed by the characterization of
its subunit 3. _Development_ 128, 4277–4288 (2001). CAS PubMed Google Scholar * Serino, G. et al. _Arabidopsis cop8_ and _fus4_ mutations define the same gene that encodes subunit 4 of
the COP9 signalosome. _Plant Cell_ 11, 1967–1980 (1999). CAS PubMed PubMed Central Google Scholar * Kwok, S. F. et al. _Arabidopsis_ homologs of a c-Jun co-activator are present both in
monomeric form and in the COP9 complex, and their abundance if differentially affected by the pleiotropic _cop/det/fus_ mutations. _Plant Cell_ 19, 1779–1790 (1998). Google Scholar *
Karniol, B., Malec, P. & Chamovitz, D. A. _Arabidopsis FUSCA5_ encodes a novel phosphoprotein that is a compoent of the COP9 complex. _Plant Cell_ 11, 839–848 (1999). CAS PubMed PubMed
Central Google Scholar * Ferrell, K., Wilkinson, C. R. M., Dubiel, W. & Gordon, C. Regulatory subunit interactions of the 26S proteasome, a complex problem. _Trends Biochem. Sci._ 25,
83–88 (2000). CAS PubMed Google Scholar * Wei, N. et al. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. _Curr. Biol._
8, 919–922 (1998). CAS PubMed Google Scholar * Peng, Z. et al. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases _in vivo_. _Curr.
Biol._ 12, R504–R505 (2003). Google Scholar * Ohi, M. D., Vander Kooi, C. W., Rosenberg, J. A., Chazin, W. J. & Gould, K. L. Structural insights into the U-box, a domain associated with
multi-ubiquitination. _Nature Struct. Biol._ 10, 250–255 (2003). CAS PubMed Google Scholar * Azevedo, C., Santos-Rosa, M. J. & Shirasu, K. The U-box protein family in plants. _Trends
Plant Sci._ 6, 354–358 (2001). CAS PubMed Google Scholar * Gagne, J. M., Downes, B. P., Shiu, S -H., Durski, A. M. & Vierstra, R. D. The F-box subunit of the SCF E3 complex is
encoded by a diverse superfamily of genes in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 99, 11519–11524 (2002). CAS PubMed PubMed Central Google Scholar * Risseeuw, E. P. et al. Protein
interaction of SCF ubiquitin E3 ligase subunits from _Arabidopsis_. _Plant J._ 34, 753–767 (2003). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS J.A.S. is supported by a
Human Frontiers Science Program long-term fellowship. Research in K.S.'s laboratory is supported in part by grants from the Gatsby Charitable Foundation and the Biotechnology and
Biological Sciences Research Council. Related research in X.W.D.'s laboratory is supported by grants from the National Institutes of Health and the National Science Foundation. AUTHOR
INFORMATION AUTHORS AND AFFILIATIONS * Deptartment of Molecular, Cellular and Developmental Biology, Yale University, PO Box 208104, 165 Prospect Street, New Haven, 06520-8104, Connecticut,
USA James A. Sullivan & Xing Wang Deng * The Sainsbury Laboratory, John Innes Centre, Colney Lane, Norwich, NR4 7UH, UK Ken Shirasu Authors * James A. Sullivan View author publications
You can also search for this author inPubMed Google Scholar * Ken Shirasu View author publications You can also search for this author inPubMed Google Scholar * Xing Wang Deng View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Xing Wang Deng. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare
that they have no competing financial interests. RELATED LINKS RELATED LINKS DATABASES TAIR _APETALA3_ _Axr1_ _CIO1_ _COP_ _COP10_ _DET_ _FUS_ _LEAFY_ _NIM1_ _PISTILLATA_ _RAR1_ _RGA_
_SGT1α_ _SGT1b_ _SKP1_ _SLEEPY1_ ubiquitin _UNUSUAL FLORAL ORGANS_ _UPL3_ UK CROPNET _Mla6_ _Mla12_ FURTHER INFORMATION Xing Wang Deng's laboratory GLOSSARY * PROTO-ONCOGENE A gene that
has the potential to change into an active cancer-causing oncogene. * ENDOCYTOSIS The uptake of extracellular materials within membrane-bound vesicles by cells. * ISOPEPTIDE BOND The
covalent linkage that joins amino-acid residues through an amide bond. * THIOESTER LINKAGE A non-covalent chemical bond between two proteins formed through thiolester. * CONJUGATION The
addition of ubiquitin moieties to a growing polyubiquitin chain. * SENESCENCE Regulated death of an organ or a cell after its normal physiological function. * CHELATION The process by which
an organic chemical bonds with metal ions and thereby removes them from solutions. * WHORLS A specific layer of flower organs that is generated through floral meristem activity. * SEPALS The
protective outer layer of flower. * STAMENS The third layer of flower that bears the male gametophyte that produces pollen. * CARPELS The fourth whorl of flower that bears the female
gametophyte. * TRANSFORMATION The change from one developmental pattern to another. * APICAL DOMINANCE Concentration of growth at the tip of a plant shoot, where a terminal bud exerts
partial inhibition of auxiliary bud growth. * ALEURONE CELLS A cell type in cereal kernals that undergoes highly regulated cell death to release stores of minerals and nutrients to the
developing embryo. * PHENOCOPY The production of a phenotype, which closely resembles a phenotype that normally results from specific gene expression or from gene mutation. * POWDERY MILDEW
A group of plant diseases that are caused by the growth of fungal mycelium and the production of spores on the surface of plant tissues. * DENEDDYLATION The removal of the ubiquitin-like
modifier NEDD8 (also called RUB) from a protein. * HISTONE DEACETYLASE The enzyme that removes acetyl groups from lysine residues in the DNA-binding histone group of proteins. *
SKOTOMORPHOGENIC Developmental pattern followed by seedlings in the absence of light. * HYPOCOTYLS The stem of a seedling. * COTYLEDONS The leaves of a seedling formed during embryonic
development. * PHOTOMORPHOGENIC Developmental pattern followed by seedlings in the presence of light. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Sullivan, J., Shirasu, K. & Deng, X. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. _Nat Rev Genet_ 4, 948–958 (2003). https://doi.org/10.1038/nrg1228
Download citation * Issue Date: 01 December 2003 * DOI: https://doi.org/10.1038/nrg1228 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative





