- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
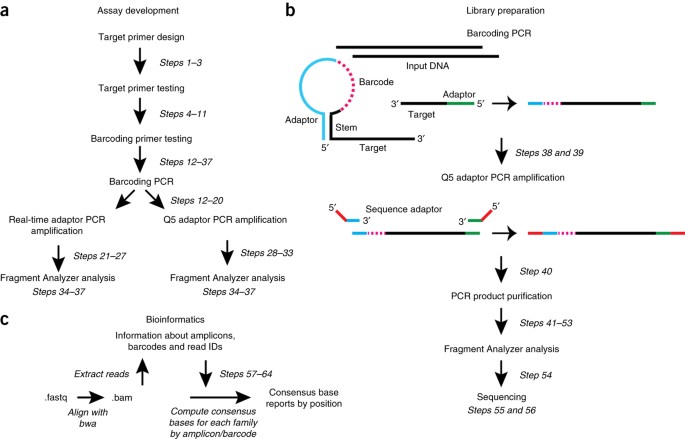
ABSTRACT Detection of extremely rare variant alleles within a complex mixture of DNA molecules is becoming increasingly relevant in many areas of clinical and basic research, such as the
detection of circulating tumor DNA in the plasma of cancer patients. Barcoding of DNA template molecules early in next-generation sequencing (NGS) library construction provides a way to
identify and bioinformatically remove polymerase errors that otherwise make detection of these rare variants very difficult. Several barcoding strategies have been reported, but all require
long and complex library preparation protocols. Simple, multiplexed, PCR-based barcoding of DNA for sensitive mutation detection using sequencing (SiMSen-seq) was developed to generate
targeted barcoded libraries with minimal DNA input, flexible target selection and a very simple, short (∼4 h) library construction protocol. The protocol comprises a three-cycle barcoding
PCR step followed directly by adaptor PCR to generate the library and then bead purification before sequencing. Thus, SiMSen-seq allows detection of variant alleles at <0.1% frequency
with easy customization of library content (from 1 to 40+ PCR amplicons) and a protocol that can be implemented in any molecular biology laboratory. Here, we provide a detailed protocol for
assay development and describe software to process the barcoded sequence reads. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel
any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SELECTIVE MULTIPLEXED ENRICHMENT FOR THE DETECTION AND QUANTITATION OF LOW-FRACTION DNA
VARIANTS VIA LOW-DEPTH SEQUENCING Article 03 May 2021 DETECTION OF LOW-FREQUENCY DNA VARIANTS BY TARGETED SEQUENCING OF THE WATSON AND CRICK STRANDS Article 03 May 2021 ADAPTOR TEMPLATE
OLIGO-MEDIATED SEQUENCING (ATOM-SEQ) IS A NEW ULTRA-SENSITIVE UMI-BASED NGS LIBRARY PREPARATION TECHNOLOGY FOR USE WITH CFDNA AND CFRNA Article Open access 04 February 2021 REFERENCES * ten
Bosch, J.R. & Grody, W.W. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. _J. Mol. Diagn._ 10, 484–492 (2008). Article CAS Google Scholar *
Fox, E.J., Reid-Bayliss, K.S., Emond, M.J. & Loeb, L.A. Accuracy of next generation sequencing platforms. _Next Gener. Seq. Appl._ 1 (2014). * Kinde, I., Wu, J., Papadopoulos, N.,
Kinzler, K.W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. _Proc. Natl. Acad. Sci. USA_ 108, 9530–9535 (2011). Article Google
Scholar * Schmitt, M.W. et al. Detection of ultra-rare mutations by next-generation sequencing. _Proc. Natl. Acad. Sci. USA_ 109, 14508–14513 (2012). Article CAS Google Scholar *
Flaherty, P. et al. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. _Nucleic Acids Res._ 40, e2 (2012). Article CAS Google Scholar * Newman, A.M.
et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. _Nat. Med._ 20, 548–554 (2014). Article CAS Google Scholar * Kennedy, S.R. et al.
Detecting ultralow-frequency mutations by duplex sequencing. _Nat. Protoc._ 9, 2586–2606 (2014). Article CAS Google Scholar * Lou, D.I. et al. High-throughput DNA sequencing errors are
reduced by orders of magnitude using circle sequencing. _Proc. Natl. Acad. Sci. USA_ 110, 19872–19877 (2013). Article CAS Google Scholar * Stahlberg, A. et al. Simple, multiplexed,
PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. _Nucleic Acids Res._ 44, e105 (2016). Article Google Scholar * Murtaza, M. et al.
Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. _Nature_ 497, 108–112 (2013). Article CAS Google Scholar * Wu, H.H. et al. Utilization of
cell-transferred cytologic smears in detection of EGFR and KRAS mutation on adenocarcinoma of lung. _Mod. Pathol._ 27, 930–935 (2013). Article Google Scholar * Boadas, J. et al. Clinical
usefulness of K-ras gene mutation detection and cytology in pancreatic juice in the diagnosis and screening of pancreatic cancer. _Eur. J. Gastroenterol. Hepatol._ 13, 1153–1159 (2001).
Article CAS Google Scholar * Ohori, N.P. et al. BRAF mutation detection in indeterminate thyroid cytology specimens: underlying cytologic, molecular, and pathologic characteristics of
papillary thyroid carcinoma. _Cancer Cytopathol._ 121, 197–205 (2013). Article CAS Google Scholar * Malapelle, U. et al. EGFR mutations detected on cytology samples by a centralized
laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. _Cancer Cytopathol._ 121, 552–560 (2013). Article CAS Google Scholar * Hoque, M.O. et al.
High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. _Cancer Res._ 63, 5723–5726 (2003). CAS PubMed
Google Scholar * Thunnissen, F.B. Sputum examination for early detection of lung cancer. _J. Clin. Pathol._ 56, 805–810 (2003). Article CAS Google Scholar * Diehl, F. et al. Analysis
of mutations in DNA isolated from plasma and stool of colorectal cancer patients. _Gastroenterology_ 135, 489–498 (2008). Article CAS Google Scholar * Forshew, T. et al. Noninvasive
identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. _Sci. Transl. Med._ 4, 136ra68 (2012). Article Google Scholar * Lo, Y.M. & Chiu, R.W.
Genomic analysis of fetal nucleic acids in maternal blood. _Annu. Rev. Genomics Hum. Genet._ 13, 285–306 (2012). Article CAS Google Scholar * New, M.I. et al. Noninvasive prenatal
diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. _J. Clin. Endocrinol. Metab._ 99, E1022–E1030 (2014). Article CAS Google Scholar * Chitty, L.S.
& Lo, Y.M. Noninvasive prenatal screening for genetic diseases using massively parallel sequencing of maternal plasma DNA. _Cold Spring Harb. Perspect. Med._ 5, a023085 (2015). Article
Google Scholar * Tsui, N.B. et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. _Blood_ 117, 3684–3691 (2011). Article CAS
Google Scholar * Li, M. et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. _Am. J. Hum. Genet._ 87, 237–249 (2010). Article CAS
Google Scholar * He, Y. et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. _Nature_ 464, 610–614 (2010). Article CAS Google Scholar * Eastman, P.S. et al.
Maternal viral genotypic zidovudine resistance and infrequent failure of zidovudine therapy to prevent perinatal transmission of human immunodeficiency virus type 1 in pediatric AIDS
Clinical Trials Group Protocol 076. _J. Infect. Dis._ 177, 557–564 (1998). Article CAS Google Scholar * McMahon, M.A. et al. The HBV drug entecavir - effects on HIV-1 replication and
resistance. _N. Engl. J. Med._ 356, 2614–2621 (2007). Article CAS Google Scholar * Snyder, T.M., Khush, K.K., Valantine, H.A. & Quake, S.R. Universal noninvasive detection of solid
organ transplant rejection. _Proc. Natl. Acad. Sci. USA_ 108, 6229–6234 (2011). Article CAS Google Scholar * Kukita, Y. et al. High-fidelity target sequencing of individual molecules
identified using barcode sequences: _de novo_ detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. _DNA Res._ 22, 269–277 (2015). Article CAS
Google Scholar * Gregory, M.T. et al. Targeted single molecule mutation detection with massively parallel sequencing. _Nucleic Acids Res._ 44, e22 (2016). Article Google Scholar *
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. _Sci. Transl. Med._ 6, 224ra24 (2014). Article Google Scholar * Shugay, M. et al.
Towards error-free profiling of immune repertoires. _Nat. Methods_ 11, 653–655 (2014). Article CAS Google Scholar * Mouliere, F. et al. High fragmentation characterizes tumour-derived
circulating DNA. _PLoS One_ 6, e23418 (2011). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the National Institutes of Health (R21CA172999 to
T.E.G.), the Swedish Cancer Society (to A.S.), the Swedish Childhood Cancer Foundation (to A.S.), the Sahlgrenska Academy at the University of Gothenburg (to A.S.). AUTHOR INFORMATION Author
notes * Anders Ståhlberg and Paul M Krzyzanowski: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Pathology and Genetics, Sahlgrenska Cancer Center,
Institute of Biomedicine, Sahlgrenska Academy at University of Gothenburg, Gothenburg, Sweden Anders Ståhlberg & Stefan Filges * Ontario Institute for Cancer Research, MaRS Centre,
Toronto, Ontario, Canada Paul M Krzyzanowski & Lincoln Stein * Department of Surgery, Boston University School of Medicine, Boston, Massachusetts, USA Matthew Egyud & Tony E Godfrey
Authors * Anders Ståhlberg View author publications You can also search for this author inPubMed Google Scholar * Paul M Krzyzanowski View author publications You can also search for this
author inPubMed Google Scholar * Matthew Egyud View author publications You can also search for this author inPubMed Google Scholar * Stefan Filges View author publications You can also
search for this author inPubMed Google Scholar * Lincoln Stein View author publications You can also search for this author inPubMed Google Scholar * Tony E Godfrey View author publications
You can also search for this author inPubMed Google Scholar CONTRIBUTIONS A.S. and T.E.G. conceived and designed the protocol; P.M.K. and L.S. developed the Debarcer software for data
analysis; and M.E. and S.F. contributed to protocol development. A.S., P.M.K., M.E., S.F. and T.E.G. wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Anders Ståhlberg or Tony E
Godfrey. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 CONCEPT OF BARCODING TO
SUPPRESS ERROR IN SEQUENCING. Each template DNA molecule is barcoded with a random and unique sequence (marked as red, turquoise and green). All PCR generated molecules that are generated
from the same original molecule receive the same barcode. If a polymerase induced error occurs (blue cross), only a fraction of all DNA molecules with the same barcode will amplify that
specific error. Conversely, a template DNA molecule with mutation will generate PCR amplicons with that particular barcode and can therefore be called a true mutant. SUPPLEMENTARY FIGURE 2
ORGANIZATION OF SIMSEN-SEQ ANALYSIS DATA PRODUCED BY DEBARCER. The Debarcer output directory contains files in the main output directory (denoted ‘Top Level’) and two subdirectories found in
this location (Tables directory and Figures directory). Files described by each unshaded box in the figure can be found in their respective directories. SUPPLEMENTARY FIGURE 3 FRAGMENT
ANALYSIS OF SINGLE-PLEX LIBRARY AT LOW DNA INPUT. Unpurified libraries were generated for a target amplicon in the _CD1C_ gene using 10, 5 and 3 ng of high quality human genomic DNA (left
side) or fragmented plasma DNA (right side) along with a no DNA control (bottom). Expected library products are indicated by the horizontal bar while all other products (<200bp) are
non-specific. Clear library PCR products can be seen in all plots except the no DNA control. SUPPLEMENTARY FIGURE 4 FRAGMENT ANALYSIS OF TRIPLEX LIBRARY AT LOW DNA INPUT. Unpurified
libraries were generated for three target amplicons in the _CD1C_, _ERBB4_, and _COL5A1_ genes using 10, 5 and 3 ng of high quality human genomic DNA (left side) or fragmented plasma DNA
(right side) along with a no DNA control (bottom). Expected library products are indicated by the horizontal bar while all other products (<200bp) are non-specific. Clear library PCR
products can be seen in all plots except the no DNA control. SUPPLEMENTARY FIGURE 5 SCHEMATIC REPRESENTATION OF THE FIRST THREE ROUNDS OF PCR USED TO INCORPORATE BARCODES. Figure shows PCR
products that are formed in cycles 1, 2 and 3 of the barcoding PCR step. Note that only DNA strands with both forward and reverse adapter sequences (or their complement) can act as templates
in the second round of PCR. These products are indicated with thicker lines. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–5 and Supplementary Table 1
(PDF 1118 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ståhlberg, A., Krzyzanowski, P., Egyud, M. _et al._ Simple multiplexed PCR-based barcoding
of DNA for ultrasensitive mutation detection by next-generation sequencing. _Nat Protoc_ 12, 664–682 (2017). https://doi.org/10.1038/nprot.2017.006 Download citation * Published: 02 March
2017 * Issue Date: April 2017 * DOI: https://doi.org/10.1038/nprot.2017.006 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative





