- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
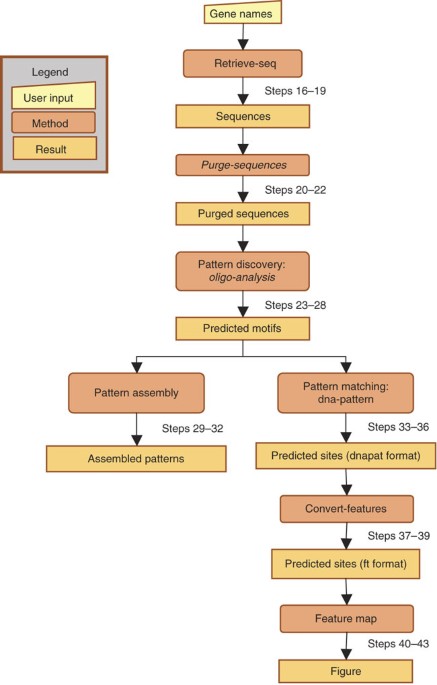
ABSTRACT This protocol shows how to access the Regulatory Sequence Analysis Tools (RSAT) via a programmatic interface in order to automate the analysis of multiple data sets. We describe the
steps for writing a Perl client that connects to the RSAT Web services and implements a workflow to discover putative _cis_-acting elements in promoters of gene clusters. In the presented
example, we apply this workflow to lists of transcription factor target genes resulting from ChIP-chip experiments. For each factor, the protocol predicts the binding motifs by detecting
significantly overrepresented hexanucleotides in the target promoters and generates a feature map that displays the positions of putative binding sites along the promoter sequences. This
protocol is addressed to bioinformaticians and biologists with programming skills (notions of Perl). Running time is ∼6 min on the example data set. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online
access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which
are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
RESTRING: AN OPEN-SOURCE PYTHON SOFTWARE TO PERFORM AUTOMATIC FUNCTIONAL ENRICHMENT RETRIEVAL, RESULTS AGGREGATION AND DATA VISUALIZATION Article Open access 06 December 2021 HOLD OUT THE
GENOME: A ROADMAP TO SOLVING THE _CIS_-REGULATORY CODE Article 13 December 2023 MASSIVELY PARALLEL CHARACTERIZATION OF INSULATOR ACTIVITY ACROSS THE GENOME Article Open access 27 September
2024 REFERENCES * Thomas-Chollier, M. et al. RSAT: regulatory sequence analysis tools. _Nucleic Acids Res._ 36, W119–W127 (2008). Article CAS Google Scholar * Brohée, S. et al. NeAT:
network analysis tools. _Nucleic Acids Res._ 36, Suppl_2 W444–W451 (2008). Article Google Scholar * Turatsinze, J.-V., Thomas-Chollier, M., Defrance, M. & van Helden, J. Using RSAT to
scan genome sequences for transcription factor binding sites and _cis_-regulatory modules. _Nat. Protoc._ doi:10.1038/nprot.2008.97 (2008). * Defrance, M., Janky, R., Sand, O. & van
Helden, J. Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. _Nat. Protoc._ doi:10.1038/nprot.2008.98 (2008). * Brohée, S., Faust, K.,
Lima-Mendez, G., Vanderstocken, G. & van Helden, J. Network Analysis Tools: from biological networks to clusters and pathways. _Nat. Protoc._ doi:10.1038/nprot.2008.100 (2008). * van
Helden, J., André, B. & Collado-Vides, J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. _J. Mol. Biol._
281, 827–842 (1998). Article CAS Google Scholar * van Helden, J. Regulatory sequence analysis tools. _Nucleic Acids Res._ 31, 3593–3596 (2003). Article CAS Google Scholar * van Helden,
J., Andre, B. & Collado-Vides, J. A web site for the computational analysis of yeast regulatory sequences. _Yeast_ 16, 177–187 (2000). Article CAS Google Scholar * Harbison, C.T. et
al. Transcriptional regulatory code of a eukaryotic genome. _Nature_ 431, 99–104 (2004). Article CAS Google Scholar * Montgomery, S.B. et al. ORegAnno: an open access database and
curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. _Bioinformatics_ 22, 637–640 (2006). Article CAS Google Scholar * Vlieghe,
D. et al. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. _Nucleic Acids Res._ 34, D95–D97 (2006). Article CAS Google Scholar *
Sandelin, A., Alkema, W., Engström, P., Wasserman, W.W. & Lenhard, B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. _Nucleic Acids Res._ 32,
D91–D94 (2004). Article CAS Google Scholar * Aerts, S. et al. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. _Nucleic Acids Res._ 33, W393–W396
(2005). Article CAS Google Scholar * Aerts, S. et al. Toucan: deciphering the cis-regulatory logic of coregulated genes. _Nucleic Acids Res._ 31, 1753–1764 (2003). Article CAS Google
Scholar * Saeed, A.I. et al. TM4: a free, open-source system for microarray data management and analysis. _Biotechniques_ 34, 374–378 (2003). Article CAS Google Scholar * Saeed, A.I. et
al. TM4 microarray software suite. _Methods Enzymol._ 411, 134–193 (2006). Article CAS Google Scholar * Reimers, M. & Carey, V.J. Bioconductor: an open source framework for
bioinformatics and computational biology. _Methods Enzymol._ 411, 119–134 (2006). Article CAS Google Scholar * van Helden, J., Rios, A.F. & Collado-Vides, J. Discovering regulatory
elements in non-coding sequences by analysis of spaced dyads. _Nucleic Acids Res._ 28, 1808–1818 (2000). Article CAS Google Scholar * Oinn, T. et al. Taverna: a tool for the composition
and enactment of bioinformatics workflows. _Bioinformatics_ 20, 3045–3054 (2004). Article CAS Google Scholar * Hull, D. et al. Taverna: a tool for building and running workflows of
services. _Nucleic Acids Res._ 34, W729–W732 (2006). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the BioSapiens Network of Excellence funded
under the sixth Framework program of the European Communities (LSHG-CT-2003-503265) (O.S. postdoc grant, E.V. research fellowship), the Vrije Universiteit Brussel (Geconcerteerde
Onderzoeksactie 29) (M.T.-C. PhD grant) and by the Belgian Program on Interuniversity Attraction Poles, initiated by the Belgian Federal Science Policy Office, project P6/25 (BioMaGNet).
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratoire de Bioinformatique des Génomes et des Réseaux (BiGRe), Université Libre de Bruxelles, Campus Plaine, CP 263, Bruxelles, Boulevard du
Triomphe, B-1050, Belgium Olivier Sand, Morgane Thomas-Chollier, Eric Vervisch & Jacques van Helden Authors * Olivier Sand View author publications You can also search for this author
inPubMed Google Scholar * Morgane Thomas-Chollier View author publications You can also search for this author inPubMed Google Scholar * Eric Vervisch View author publications You can also
search for this author inPubMed Google Scholar * Jacques van Helden View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence
to Jacques van Helden. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 Full version of the feature map showing the positions of the significant hexanucleotides found in promoters of genes
from the BAS1_YPD cluster. (PDF 82 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Sand, O., Thomas-Chollier, M., Vervisch, E. _et al._ Analyzing
multiple data sets by interconnecting RSAT programs via SOAP Web services—an example with ChIP-chip data. _Nat Protoc_ 3, 1604–1615 (2008). https://doi.org/10.1038/nprot.2008.99 Download
citation * Published: 18 September 2008 * Issue Date: October 2008 * DOI: https://doi.org/10.1038/nprot.2008.99 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative

![[withdrawn] yjb to withdraw from hassockfield stc and hindley yoi](https://assets.publishing.service.gov.uk/media/5a62111ee5274a0a3ad908a5/s960_YJB_logo_Gov.uk.jpg)

![[withdrawn] aggregate industries uk limited: application made to abstract water](https://www.gov.uk/assets/static/govuk-opengraph-image-03837e1cec82f217cf32514635a13c879b8c400ae3b1c207c5744411658c7635.png)
