- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
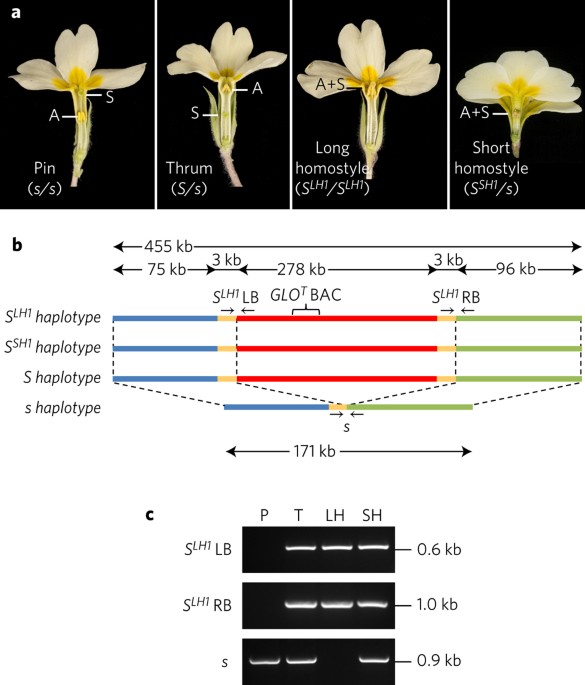
ABSTRACT Darwin's studies on heterostyly in _Primula_ described two floral morphs, pin and thrum, with reciprocal anther and stigma heights that promote insect-mediated
cross-pollination. This key innovation evolved independently in several angiosperm families. Subsequent studies on heterostyly in _Primula_ contributed to the foundation of modern genetic
theory and the neo-Darwinian synthesis. The established genetic model for _Primula_ heterostyly involves a diallelic _S_ locus comprising several genes, with rare recombination events that
result in self-fertile homostyle flowers with anthers and stigma at the same height. Here we reveal the _S_ locus supergene as a tightly linked cluster of thrum-specific genes that are
absent in pins. We show that thrums are hemizygous not heterozygous for the _S_ locus, which suggests that homostyles do not arise by recombination between _S_ locus haplotypes as previously
proposed. Duplication of a floral homeotic gene 51.7 million years (Myr) ago, followed by its neofunctionalization, created the current _S_ locus assemblage which led to floral heteromorphy
in _Primula._ Our findings provide new insights into the structure, function and evolution of this archetypal supergene. Access through your institution Buy or subscribe This is a preview
of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and online access to articles $119.00
per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS CONVERGENT
EVOLUTIONARY PATTERNS OF HETEROSTYLY ACROSS ANGIOSPERMS SUPPORT THE POLLINATION-PRECISION HYPOTHESIS Article Open access 09 February 2024 COMPARATIVE TRANSCRIPTOMICS REVEALS COMMONALITIES
AND DIFFERENCES IN THE GENETIC UNDERPINNINGS OF A FLORAL DIMORPHISM Article Open access 01 December 2022 GENOME OF THE EARLY SPIDER-ORCHID _OPHRYS SPHEGODES_ PROVIDES INSIGHTS INTO SEXUAL
DECEPTION AND POLLINATOR ADAPTATION Article Open access 26 July 2024 REFERENCES * Barrett, S. C. H. The evolution of plant sexual diversity. _Nat. Rev. Genet._ 3, 274–284 (2002). Article
CAS Google Scholar * Richards, A. J. _Primula_ 2nd edn (Batsford, 2002). Google Scholar * Darwin, C. R. On the two forms or dimorphic condition in the species of _Primula_, and on their
remarkable sexual relations. _J. Proc. Linn. Soc. Bot._ 6, 77–96 (1862). Google Scholar * Gregory, R. P., De Winton, D. & Bateson, M. A. Genetics of _Primula sinensis_. _J. Genet._ 13,
219–253 (1923). Article Google Scholar * Bateson, W. & Gregory, R. P. On the inheritance of heterostylism in _Primula_. _Proc. R. Soc. Lond. B_ 76, 581–586 (1905). Article Google
Scholar * Bridges, C. B. The chromosome hypothesis of linkage applied to cases in sweetpeas and _Primula_. _Am. Nat._ 48, 524–534 (1914). Article Google Scholar * Ernst, A. Weitere
untersuchungen zur phänanalyse zum fertilitätsproblem und zur genetik heterostyler primeln. II. Primula hortensis. _Arch. Julius Klaus Stift. Vererbungsforsch. Sozialanthropol. Rassenhyg._
11, 1–280 (1936). Google Scholar * Ernst, A. Heterostylie-Forschung versuche zur genetischen analyse eines organisations und ‘Anpassungs’ merkmales. _Z. Induk. Abstamm. Vererbungsl._ 71,
156–230 (1936). Google Scholar * De Winton, D. & Haldane, J. B. S. The genetics of _Primula sinensis_. III. Linkage in the diploid. _J. Genet._ 31, 67–100 (1935). Article Google
Scholar * Darlington, C. D. Meiosis in diploid and tetraploid _Primula sinensis_. _J. Genet._ 24, 65–95 (1931). Article Google Scholar * Mather, K. The genetical architecture of
heterostyly in _Primula sinensis_. _Evolution_ 4, 340–352 (1950). Article Google Scholar * Schwander, T., Libbrecht, R. & Keller, L. Supergenes and complex phenotypes. _Curr. Biol._
24, R288–R294 (2014). Article CAS Google Scholar * Darwin, C. R. _The Different Forms of Flowers on Plants of the Same Species_ (John Murray, 1877). Book Google Scholar * Dodd, M. E.,
Silvertown, J. & Chase, M. W. Phylogenetic analysis of trait evolution and species diversity variation among angiosperm families. _Evolution_ 53, 732–744 (1999). Article Google Scholar
* Webster, M. A. & Gilmartin, P. M. Analysis of late stage flower development in primula vulgaris reveals novel differences in cell morphology and temporal aspects of floral
heteromorphy. _New Phytol._ 171, 591–603 (2006). PubMed Google Scholar * Shivanna, K. R., Heslop-Harrison, J. & Heslop-Harrison, Y. Heterostyly in primula. 2. Sites of pollen
inhibition, and effects of pistil constituents on compatible and incompatible pollen tube growth. _Protoplasma_ 107, 319–337 (1981). Article Google Scholar * Richards, A. J. & Ibrahim,
H. B. The breeding system in primula veris L .II. pollen-Tube growth and seed-Set. _New Phytol._ 90, 305–314 (1982). Article Google Scholar * Lewis, D. Comparative incompatibility in
angiosperms and fungi. _Adv. Genet._ 6, 235–285 (1954). Article CAS Google Scholar * Dowrick, V. P. J. Heterostyly and homostyly in _Primula obconica_. _Heredity_ 10, 219–236 (1956).
Article Google Scholar * Lloyd, D. G. & Webb, C. J. in _Evolution and Function of Heterostyly_ (ed. Barrett, S. C. H. ) 151–175 (Springer Verlag, 1992). Book Google Scholar *
Charlesworth, D. & Charlesworth, B. Model for the evolution of distyly. _Am. Nat._ 114, 467–498 (1979). Article Google Scholar * Bodmer, W. F. The genetics of homostyly in populations
of _Primula vulgaris_. _Phil. Trans. R. Soc. Lond. B_ 242, 517–549 (1960). Article Google Scholar * Fisher, R. A. A theoretical system of selection for homostyle. _Primula Sankhya._ 9,
325–342 (1949). Google Scholar * Piper, J. G., Charlesworth, B. & Charlesworth, D. A high-rate of self-fertilization and increased seed fertility of homostyle primroses. _Nature._ 310,
50–51 (1984). Article Google Scholar * Crosby, J. L. High proportions of homostyle plants in populations of _Primula vulgaris_. _Nature._ 145, 672–673 (1940). Article Google Scholar *
Crosby, J. L. Selection of an unfavourable gene complex. _Evol. Ecol. Res._ 3, 212–230 (1949). CAS Google Scholar * Webster, M. A. & Gilmartin, P. M. A comparison of early floral
ontogeny in wild-type and floral homeotic mutant phenotypes of _Primula. Planta_ 216, 903–917 (2003). CAS PubMed Google Scholar * McCubbin, A. G., Lee, C. & Hetrick, A. Identification
of genes showing differential expression between morphs in developing flowers of _Primula vulgaris_. _Sex. Plant Reprod._ 19, 63–72 (2006). Article CAS Google Scholar * Li, J., Webster,
M. A., Furuya, M. & Gilmartin, P. M. Identification and characterization of pin and thrum alleles of two genes that co-segregate with the _Primula S_ locus. _Plant J._ 51, 18–31 (2007).
Article CAS Google Scholar * Manfield, I. W. _et al._ Molecular characterization of DNA sequences from the _Primula vulgaris_ _S_ locus. _J. Exp. Bot._ 56, 1177–1188 (2005). Article CAS
Google Scholar * Cocker, J. _et al._ Oakleaf: an S locus-linked mutation of _Primula vulgaris_ that affects leaf and flower development. _New Phytol._ 208, 149–161 (2015). Article CAS
Google Scholar * Li, J. _et al._ _Hose in Hose_, an _S_ locus-linked mutant of _Primula vulgaris_ is caused by an unstable mutation at the _Globosa_ locus. _Proc. Natl Acad. Sci. USA_ 107,
5664–5668 (2010). Article CAS Google Scholar * Li, J. _et al._ The _S_ locus-linked _Primula_ homeotic mutant _sepaloid_ shows characteristics of a B-function mutant but does not result
from mutation in a B-function gene. _Plant J._ 56, 1–12 (2008). Article CAS Google Scholar * Yoshida, Y. _et al._ QTL analysis of heterostyly in _Primula sieboldii_ and its application
for morph identification in wild populations. _Ann. Bot._ 108, 133–142 (2011). Article Google Scholar * Li, J. _et al_. Integration of genetic and physical maps of the _Primula vulgaris S_
locus and localization by chromosome _in situ_ hybridisation. _New Phytol._ 208, 137–148 (2015). Article CAS Google Scholar * Nowak, M. D. _et al._ The draft genome of _Primula veris_
yields insight into the molecular basis of heterostyly. _Genome Biol._ 16, 16 (2015). Article Google Scholar * Verhoef, N. _et al._ Brassinosteroid biosynthesis and signalling in _Petunia
hybrida_. _J. Exp. Bot._ 64, 2435–2448 (2013). Article CAS Google Scholar * Turk, E. M. _et al._ CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis
and plant steroid signal transduction. _Plant Physiol._ 133, 1643–1653 (2003). Article CAS Google Scholar * Abbasi, N., Park, Y.-I. & Choi, S.-B. Pumilio Puf domain RNA-binding
proteins in _Arabidopsis_. _Plant Signal. Behav._ 6, 364–368 (2011). Article CAS Google Scholar * Kim, H. J., Chiang, Y.-H., Kieber, J. J. & Schaller, G. E. SCFKMD controls cytokinin
signaling by regulating the degradation of type-B response regulators. _Proc. Natl Acad. Sci. USA_ 110, 10028–10033 (2013). Article CAS Google Scholar * Webster, M. A. & Grant, C. J.
The inheritance of calyx morph variants in _Primula vulgaris_ (Huds). _Heredity_ 64, 121–124 (1990). Article Google Scholar * Viaene, T. _et al._ Pistillata-duplications as a mode for
floral diversification in (Basal) asterids. _Mol. Biol. Evol._ 26, 2627–2645 (2009). Article CAS Google Scholar * Xia, X. DAMBE5: a comprehensive software package for data analysis in
molecular biology and evolution. _Mol. Biol. Evol._ 30, 1720–1728 (2013). Article CAS Google Scholar * Magallón, S., Gómez-Acevedo, S., Sánchez-Reyes, L. L. & Hernández-Hernández, T.
A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. _New Phytol._ 207, 437–453 (2015). Article Google Scholar * Bell, C. D., Soltis, D. E. &
Soltis, P. S. The age and diversification of the angiosperms re-revisited. _Am. J. Bot._ 97, 1296–1303 (2010). Article Google Scholar * Mast, A. R. _et al._ Phylogenetic relationships in
_Primula_ L. and related genera (Primulaceae) based on noncoding chloroplast DNA. _Int. J. Plant Sci._ 162, 1381–1400 (2001). Article CAS Google Scholar * Joron, M. _et al._ Chromosomal
rearrangements maintain a polymorphic supergene controlling butterfly mimicry. _Nature_ 477, 203–206 (2011). Article CAS Google Scholar * Thomas, J. W. _et al._ The chromosomal
polymorphism linked to variation in social behavior in the white-throated sparrow (_Zonotrichia albicollis_) is a complex rearrangement and suppressor of recombination. _Genetics_ 179,
1455–1468 (2008). Article CAS Google Scholar * Wang, J. _et al._ A Y-like social chromosome causes alternative colony organization in fire ants. _Nature_ 493, 664–668 (2013). Article CAS
Google Scholar * Turgeon, B. G. & Yoder, O. C. Proposed nomenclature for mating type genes of filamentous ascomycetes. _Fungal Genet. Biol._ 31, 1–5 (2000). Article CAS Google
Scholar Download references ACKNOWLEDGEMENTS We thank M. Lappage, M. Hughes and P. Wells for horticultural support; colleagues at TGAC for Illumina sequencing; A. Thanki for TGAC Browser
support; O. Kent for _P. elatior_ _GLO_ and _GLO__T_ sequences; Norfolk Wildlife Trust, Suffolk Wildlife Trust and Norfolk County Council for permission to sample _P. veris_, _P. elatior_
and _P. vulgaris_ respectively; M. Gage, B. Davies and D. Bowles for comments on the manuscript; W. Wang for advice on _k_-means analysis; BBSRC for funding via grant BB/H019278/2, and prior
awards G11027 and P11021; The Gatsby Foundation for early stage funding; University of Leeds, Durham University and University of East Anglia for support to P.M.G. over several years of the
project. CvO was funded by the Earth & Life Systems Alliance (ELSA). P.M.G.'s laboratory is hosted at the John Innes Centre under the UEA-JIC Norwich Research Park collaboration.
AUTHOR INFORMATION Author notes * Sarah Dyer & Mario Caccamo Present address: †Present address: National Institute for Agricultural Botany, Huntingdon Road, Cambridge CB3 0LE, UK, *
Jinhong Li and Jonathan M. Cocker: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * School of Biological Sciences, University of East Anglia, Norwich Research Park,
Norwich, NR4 7TJ, UK Jinhong Li, Jonathan M. Cocker, Margaret A. Webster & Philip M. Gilmartin * John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK Jinhong Li, Jonathan M.
Cocker, Margaret A. Webster & Philip M. Gilmartin * The Earlham Institute, Norwich Research Park, Norwich, NR4 7UH, UK Jonathan Wright, Mark McMullan, Sarah Dyer, David Swarbreck &
Mario Caccamo * School of Environmental Sciences, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK Cock van Oosterhout Authors * Jinhong Li View author publications You
can also search for this author inPubMed Google Scholar * Jonathan M. Cocker View author publications You can also search for this author inPubMed Google Scholar * Jonathan Wright View
author publications You can also search for this author inPubMed Google Scholar * Margaret A. Webster View author publications You can also search for this author inPubMed Google Scholar *
Mark McMullan View author publications You can also search for this author inPubMed Google Scholar * Sarah Dyer View author publications You can also search for this author inPubMed Google
Scholar * David Swarbreck View author publications You can also search for this author inPubMed Google Scholar * Mario Caccamo View author publications You can also search for this author
inPubMed Google Scholar * Cock van Oosterhout View author publications You can also search for this author inPubMed Google Scholar * Philip M. Gilmartin View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS J.L. contributed to project design, performed all molecular analyses, generated the _S_ locus assembly, manually annotated the
_S_ locus gene structures and undertook data analysis. J.M.C. carried out bioinformatic analyses, including automated annotation of the _S_ locus region, undertook _in silico_ gene
expression and _k_-means clustering analyses, assembled genome sequences and library scaffolds, generated the molecular phylogeny, undertook recombination analysis of the _S_ locus flanking
regions and contributed to project design. J.W. assembled genome sequences and library scaffolds, contributed to genome annotation and generated the automated gene model predictions across
the _S_ locus, aligned sequencing reads to the _S_ locus assembly and contributed to project design. M.A.W. contributed the inbred long homostyle line, other genetic resources and classical
genetics, identified the short homostyle mutant and generated the three-point cross used to demonstrate linkage. M.M. and C.v.O. contributed to the molecular phylogeny construction,
evolutionary data analysis and recombination analysis. S.A., D.S. and M.C. contributed to the genome sequencing strategy, assembly and annotation that underpins this project. P.M.G.
conceived, designed and directed the project, contributed to data analysis, prepared the figures and drafted the manuscript, with revision input from C.v.O.; all authors contributed to
editing the manuscript. CORRESPONDING AUTHOR Correspondence to Philip M. Gilmartin. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Methods, Supplementary References, Supplementary Figures 1–4, Supplementary Tables 1–6, Supplementary Sequence Analyses 1–3.
(PDF 1701 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, J., Cocker, J., Wright, J. _et al._ Genetic architecture and evolution of the _S_
locus supergene in _Primula vulgaris_. _Nature Plants_ 2, 16188 (2016). https://doi.org/10.1038/nplants.2016.188 Download citation * Received: 08 September 2015 * Accepted: 31 October 2016 *
Published: 02 December 2016 * DOI: https://doi.org/10.1038/nplants.2016.188 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative







