- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
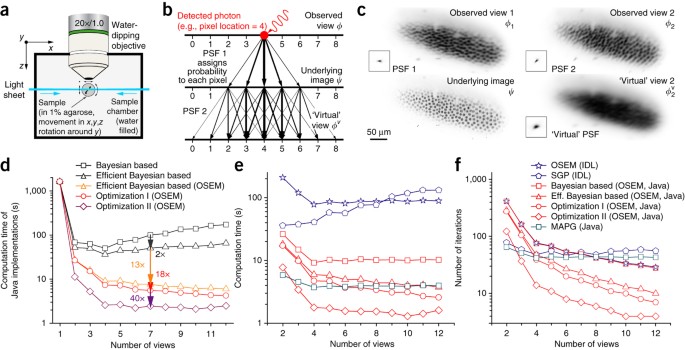
ABSTRACT Light-sheet fluorescence microscopy is able to image large specimens with high resolution by capturing the samples from multiple angles. Multiview deconvolution can substantially
improve the resolution and contrast of the images, but its application has been limited owing to the large size of the data sets. Here we present a Bayesian-based derivation of multiview
deconvolution that drastically improves the convergence time, and we provide a fast implementation using graphics hardware. Access through your institution Buy or subscribe This is a preview
of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during
checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS APPROACHING MAXIMUM
RESOLUTION IN STRUCTURED ILLUMINATION MICROSCOPY VIA ACCURATE NOISE MODELING Article Open access 31 January 2025 EXTENDING RESOLUTION WITHIN A SINGLE IMAGING FRAME Article Open access 02
December 2022 SPARSE DECONVOLUTION IMPROVES THE RESOLUTION OF LIVE-CELL SUPER-RESOLUTION FLUORESCENCE MICROSCOPY Article 15 November 2021 REFERENCES * Huisken, J., Swoger, J., Del Bene, F.,
Wittbrodt, J. & Stelzer, E.H.K. _Science_ 305, 1007–1009 (2004). Article CAS Google Scholar * Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. _Science_ 322, 1065–1069
(2008). Article CAS Google Scholar * Truong, T.V., Supatto, W., Koos, D.S., Choi, J.M. & Fraser, S.E. _Nat. Methods_ 8, 757–760 (2011). Article CAS Google Scholar * Swoger, J.,
Verveer, P., Greger, K., Huisken, J. & Stelzer, E.H.K. _Opt. Express_ 15, 8029–8042 (2007). Article Google Scholar * Shepp, L.A. & Vardi, Y. _IEEE Trans. Med. Imaging_ 1, 113–122
(1982). Article CAS Google Scholar * Hudson, H.M. & Larkin, R.S. _IEEE Trans. Med. Imaging_ 13, 601–609 (1994). Article CAS Google Scholar * Verveer, P.J. et al. _Nat. Methods_ 4,
311–313 (2007). Article CAS Google Scholar * Bonettini, S., Zanella, R. & Zanni, L. _Inverse Probl._ 25, 015002 (2009). Article Google Scholar * Krzic, U. _Multiple-View Microscopy
with Light-Sheet Based Fluorescent Microscope_. PhD thesis, Univ. Heidelberg (2009). * Temerinac-Ott, M. et al. _IEEE Trans. Image Process._ 21, 1863–1873 (2012). Article Google Scholar *
Richardson, W.H. _J. Opt. Soc. Am._ 62, 55–59 (1972). Article Google Scholar * Lucy, L.B. _Astron. J._ 79, 745–754 (1974). Article Google Scholar * Dempster, A.P., Laird, N.M. &
Rubin, D.B. _J. R. Stat. Soc. Series B Stat. Methodol._ 39, 1–38 (1977). Google Scholar * Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. _Nat. Methods_ 7, 418–419 (2010).
Article CAS Google Scholar * Tikhonov, A.N. & Arsenin, V.Y. _Solutions of Ill-Posed Problems_ (Winston, 1977). * Long, F., Peng, H., Liu, X., Kim, S. & Myers, E. _Nat. Methods_ 6,
667–672 (2009). Article CAS Google Scholar * Pitrone, P.G. et al. _Nat. Methods_ 10, 598–599 (2013). Article CAS Google Scholar * Keller, P.J. et al. _Nat. Methods_ 7, 637–642 (2010).
Article CAS Google Scholar * Schindelin, J. et al. _Nat. Methods_ 9, 676–682 (2012). Article CAS Google Scholar * Pietzsch, T., Preibisch, S., Tomancak, P. & Saalfeld, S.
_Bioinformatics_ 28, 3009–3011 (2012). Article CAS Google Scholar * Uddin, M.S., Lee, H.K., Preibisch, S. & Tomancak, P. _Microsc. Microanal._ 17, 607–613 (2011). Article CAS Google
Scholar Download references ACKNOWLEDGEMENTS We thank T. Pietzsch (Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG)) for helpful discussions, proofreading and access
to his unpublished software; N. Clack, F. Carrillo Oesterreich and H. Bowne-Anderson for discussions; N. Maghelli for two-photon imaging; P. Verveer (MPI Dortmund) for source code and
helpful discussions; M. Weber for imaging the _Drosophila_ time series; S. Jaensch for preparing the _C. elegans_ embryo; J.K. Liu (Cornell University) for the LW698 strain; S. Saalfeld for
help with 3D rendering; P.J. Keller for supporting F.A. and for the DSLM-SI data set; A. Cardona for access to his computer; and Carl Zeiss Microimaging for providing us with the SPIM
prototype. S.P. was supported by MPI-CBG in P.T.'s lab, Howard Hughes Medical Institute (HHMI) in E.M.'s lab and the Human Frontier Science Program (HFSP) Postdoctoral Fellowship
LT000783/2012 in R.H.S.'s lab, with additional support from US National Institutes of Health (NIH) GM57071. F.A. was supported by HHMI in P.J. Keller's lab. E.S. and M.S. were
supported by MPI-CBG. R.H.S. was supported by NIH grants GM057071, EB013571 and NS083085. E.M. was supported by HHMI and MPI-CBG. P.T. was supported by The European Research Council
Community's Seventh Framework Program (FP7/2007-2013) grant agreement 260746 and the HFSP Young Investigator grant RGY0093/2012. M.S., E.M. and P.T. were additionally supported by the
Bundesministerium für Bildung und Forschung grant 031A099. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany Stephan
Preibisch, Evangelia Stamataki, Mihail Sarov, Eugene Myers & Pavel Tomancak * Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, Virginia, USA Stephan Preibisch,
Fernando Amat, Robert H Singer & Eugene Myers * Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, Bronx, New York, USA Stephan Preibisch & Robert H
Singer * Gruss Lipper Biophotonics Center, Albert Einstein College of Medicine, Bronx, New York, USA Stephan Preibisch & Robert H Singer Authors * Stephan Preibisch View author
publications You can also search for this author inPubMed Google Scholar * Fernando Amat View author publications You can also search for this author inPubMed Google Scholar * Evangelia
Stamataki View author publications You can also search for this author inPubMed Google Scholar * Mihail Sarov View author publications You can also search for this author inPubMed Google
Scholar * Robert H Singer View author publications You can also search for this author inPubMed Google Scholar * Eugene Myers View author publications You can also search for this author
inPubMed Google Scholar * Pavel Tomancak View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS S.P. and F.A. derived the equations for multiview
deconvolution. S.P. implemented the software and performed all analysis, and F.A. implemented the GPU code. E.S. generated and imaged the H2Av-mRFPruby fly line. M.S. prepared, and M.S. and
S.P. imaged, the _C. elegans_ L1 sample. S.P. and P.T. conceived the idea and wrote the manuscript. R.H.S. provided support and encouragement, E.M. and P.T. supervised the project.
CORRESPONDING AUTHORS Correspondence to Stephan Preibisch or Pavel Tomancak. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–17, Supplementary Table 1 and Supplementary Notes 1–8 (PDF 12761 kb) VISUALIZATION OF THE MULTIVIEW DECONVOLUTION PROCESS
ON SYNTHETIC DATA The movie illustrates the core idea underlying the multiview deconvolution process. The deconvolution is finished once each view is identical to the deconvolved image
convolved with the respective PSF, i.e. the deconvolved image together with the PSF's completely explains each observed view. To achieve this, the deconvolution first computes the
difference between the input image and the convolution of the current guess of the underlying image with the PSF. Based on this difference each view suggests an update that is then combined
into one update of the deconvolved image. In this movie the iterations continue until a cross correlation of 0.99 is achieved. For this synthetic data picturing lines, 21944 iterations were
required. (MOV 1264 kb) DECONVOLUTION OF SIMULATED DATASET (LATERAL ORIENTATION, XY) Shows all slices in lateral orientation of view 1 through the simulated ground truth, view 1,
content-based fused image, MAPG deconvolution, Optimization II deconvolution and Optimization II deconvolution with Tikhonov regularization (lamdba = 0.004). (MOV 3200 kb) DECONVOLUTION OF
SIMULATED DATASET (ORIENTATION ALONG ROTATION AXIS, YZ) Shows all slices oriented along the rotation axis of the virtual microscope. Cut planes are shown through the simulated ground truth,
view 1, content-based fused image, MAPG deconvolution, Optimization II deconvolution and Optimization II deconvolution with Tikhonov regularization (lamdba = 0.004). (MOV 3235 kb) FIXED L1
_C. ELEGANS_ LARVAE ORIENTATION 1 Fixed _C. elegans_ larvae in L1 stage expressing LMN-1::GFP (green) and stained with Hoechst (magenta) imaged on Zeiss Lightsheet Z1 (Carl Zeiss
Microimaging) with a 20x/1.0 detection objective using four views (0, 45, 90, 135 degrees). The specimen is oriented so that the required bounding box is minimized. The top panel shows a
stack of input view 0 close to its original orientation; the lower panel shows the corresponding deconvolved stack. (MOV 2035 kb) FIXED L1 _C. ELEGANS_ LARVAE ORIENTATION 2 Fixed _C.
elegans_ larvae in L1 stage expressing LMN-1::GFP (green) and stained with Hoechst (magenta) imaged on Zeiss Lightsheet Z.1 (Carl Zeiss Microimaging) with a 20x/1.0 detection objective using
four views (0, 45, 90, 135 degrees). The specimen is oriented so that the required bounding box is minimized. The top view pictures the stack of input view 0 in an almost axial orientation
(orthogonal to SUPPLEMENTARY VIDEO 4), the lower panel shows the corresponding deconvolved stack. (MOV 1983 kb) FIXED L1 _C. ELEGANS_ LARVAE ALONG ROTATION AXIS Fixed _C. elegans_ larvae in
L1 stage expressing LMN-1::GFP (green) and stained with Hoechst (magenta) imaged on Zeiss Lightsheet Z1 (Carl Zeiss Microimaging) with a 20x/1.0 detection objective using four views (0, 45,
90, 135 degrees). The specimen is oriented so that the required bounding box is minimized. The top part pictures the stack of input view 0 approximately along the rotation axis of the
microscope (orthogonal to SUPPLEMENTARY VIDEO 4 & 5), the lower panel shows the corresponding deconvolved stack. Note that none of the input views offers high resolution in this
orientation. (MOV 2821 kb) SEGMENTATION OF FIXED L1 _C. ELEGANS_ LARVAE 3D maximal projection of the manual segmentation of the deconvolved fixed _C. elegans_ larvae in L1 stage expressing
LMN-1::GFP and stained with Hoechst. 558 nuclei were manually identified using the ImgLib219 based viewing and annotation software developed by Tobias Pietzsch (unpublished software). (MOV
3565 kb) DECONVOLUTION OF _DROSOPHILA_ EMBRYOGENESIS TIME AND SERIES ORIENTATION 1 _Drosophila melanogaster_ embryo expressing His-YFP in all nuclei imaged on a Zeiss SPIM prototype with a
20x/0.5NA detection objective using 7 views. The top panel shows a 3d rendering of the side view of the deconvolved volume, the lower panel the result of the content-based fusion. (MOV 6223
kb) DECONVOLUTION OF _DROSOPHILA_ EMBRYOGENESIS TIME AND SERIES ORIENTATION 2 _Drosophila melanogaster_ embryo expressing His-YFP in all nuclei imaged on a Zeiss SPIM prototype with a
20x/0.5NA detection objective using 7 views. The top panel shows a 3d rendering of dorsal side of the deconvolved volume, the lower panel the result of the content-based fusion. (MOV 6006
kb) DECONVOLUTION OF _DROSOPHILA_ EMBRYOGENESIS TIME AND SERIES VIEW ALONG ROTATION AXIS _Drosophila melanogaster_ embryo expressing His-YFP in all nuclei imaged on a Zeiss SPIM prototype
with a 20x/0.5NA detection objective using 7 views. The top panel shows a 3d rendering of the anterior part, the lower panel of the posterior part of the embryo. The left side pictures the
deconvolved volume, on the right side the result of the content-based fusion. Note that these renderings of the sample reconstitute a view along the rotation axis of the microscope, i.e. no
views were acquired by the microscope in this orientation and therefore the increase in resolution by the deconvolution is maximal. (MOV 6549 kb) DECONVOLUTION OF _DROSOPHILA_ EMBRYOGENESIS
TIME AND SERIES ACQUIRED WITH LIGHTSHEET Z1 _Drosophila melanogaster_ embryo expressing Histone-H2Av-mRFPruby fusion in all cells was imaged on Zeiss Lightsheet Z1 (Carl Zeiss Microimaging)
with a 20x/1.0 detection objective using dual sided illumination from six angles every 1.5 minutes for 17 hours (715 timepoints). The recording starts right before initiation of gastrulation
movements and ends when the embryo hatches into a first instar larva and crawls out of the imaged volume. The data were registered using Fiji's bead based multiview SPIM registration
plugin15. The two times downsampled registered data were fused into a single output image using content based fusion (top) and multiview deconvolution with 10 iterations and Tikhonov
regularization parameter set to 0.0006 (bottom). The fused and deconvolved data were independently rendered using Fiji/ImgLib219 based cpu renderer (Stephan Saalfeld unpublished software).
All processing was performed on a cluster computer using Fiji launched with custom shell scripts (http://fiji.sc/SPIM_Registration_on_cluster). The deconvolution took advantage of four nodes
with NVIDIA Tesla graphics card and ran for the 715 time points for about 35 hours (i.e 4 more graphics cards would make the processing real time, 17 hours of processing for ∼17 hour
acquisition). (MOV 33862 kb) SUPPLEMENTARY SOFTWARE 1 Software for the deconvolution of multiview microscopy data (Fiji plugin) (ZIP 361 kb) SUPPLEMENTARY SOFTWARE 2 Software simulating
multiview selective plane microscopy data including signal attenuation, convolution, reduced axial sampling and Poisson noise. (ZIP 421 kb) SUPPLEMENTARY SOFTWARE 3 CUDA implementation of
the Fourier convolution for fast processing (additional library for Fiji plugin) (ZIP 66 kb) SOURCE DATA SOURCE DATA TO FIG. 1 SOURCE DATA TO FIG. 2 SOURCE DATA TO FIG. 3 RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Preibisch, S., Amat, F., Stamataki, E. _et al._ Efficient Bayesian-based multiview deconvolution. _Nat Methods_ 11,
645–648 (2014). https://doi.org/10.1038/nmeth.2929 Download citation * Received: 08 July 2013 * Accepted: 06 March 2014 * Published: 20 April 2014 * Issue Date: June 2014 * DOI:
https://doi.org/10.1038/nmeth.2929 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative







