- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
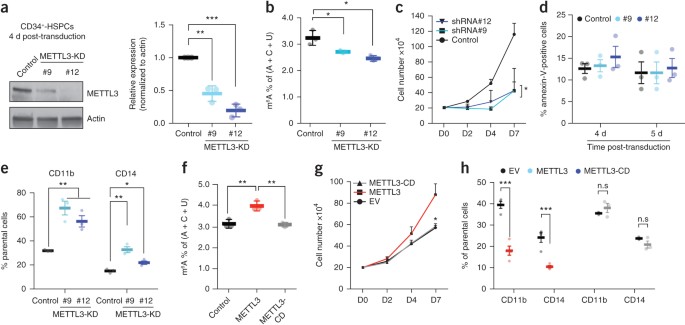
ABSTRACT _N_6-methyladenosine (m6A) is an abundant nucleotide modification in mRNA that is required for the differentiation of mouse embryonic stem cells. However, it remains unknown whether
the m6A modification controls the differentiation of normal and/or malignant myeloid hematopoietic cells. Here we show that shRNA-mediated depletion of the m6A-forming enzyme METTL3 in
human hematopoietic stem/progenitor cells (HSPCs) promotes cell differentiation, coupled with reduced cell proliferation. Conversely, overexpression of wild-type METTL3, but not of a
catalytically inactive form of METTL3, inhibits cell differentiation and increases cell growth. _METTL3_ mRNA and protein are expressed more abundantly in acute myeloid leukemia (AML) cells
than in healthy HSPCs or other types of tumor cells. Furthermore, METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia
progression in recipient mice _in vivo_. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of _c-MYC_, _BCL2_ and _PTEN_
mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT, which contributes to the differentiation-promoting
effects of METTL3 depletion. Overall, these results provide a rationale for the therapeutic targeting of METTL3 in myeloid leukemia. Access through your institution Buy or subscribe This is
a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value
online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more
Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS:
* Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS THE M6A READER IGF2BP3 PROMOTES ACUTE MYELOID LEUKEMIA
PROGRESSION BY ENHANCING RCC2 STABILITY Article Open access 25 February 2022 NEW INSIGHT INTO THE CATALYTIC -DEPENDENT AND -INDEPENDENT ROLES OF METTL3 IN SUSTAINING ABERRANT TRANSLATION IN
CHRONIC MYELOID LEUKEMIA Article Open access 24 September 2021 YTHDC1-MEDIATED MICRORNA MATURATION IS ESSENTIAL FOR HEMATOPOIETIC STEM CELLS MAINTENANCE Article Open access 16 October 2024
ACCESSION CODES ACCESSIONS GENE EXPRESSION OMNIBUS * GSE98623 REFERENCES * Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons.
_Cell_ 149, 1635–1646 (2012). Article CAS Google Scholar * Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. _Nature_ 485, 201–206 (2012).
Article CAS Google Scholar * Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. _Science_ 347, 1002–1006 (2015).
Article CAS Google Scholar * Batista, P.J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. _Cell Stem Cell_ 15, 707–719 (2014). Article CAS
Google Scholar * Li, Z. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. _Cancer Cell_ 31, 127–141 (2017). Article Google Scholar *
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. _Nature_ 541, 371–375 (2017). Article CAS Google Scholar * Wang, X. et al. Corrigendum: Structural
basis of N6-adenosine methylation by the METTL3-METTL14 complex. _Nature_ 542, 260 (2017). Article CAS Google Scholar * Lin, S., Choe, J., Du, P., Triboulet, R. & Gregory, R.I. The
m6A methyltransferase METTL3 promotes translation in human cancer cells. _Mol. Cell_ 62, 335–345 (2016). Article CAS Google Scholar * Bagger, F.O. et al. BloodSpot: a database of gene
expression profiles and transcriptional programs for healthy and malignant haematopoiesis. _Nucleic Acids Res._ 44 D1, D917–D924 (2016). Article CAS Google Scholar * Śledź, P. &
Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. _eLife_ 5, e18434 (2016). Article Google Scholar * Wang, P., Doxtader, K.A. & Nam, Y. Structural
basis for cooperative function of Mettl3 and Mettl14 methyltransferases. _Mol. Cell_ 63, 306–317 (2016). Article CAS Google Scholar * Wang, X. et al. Structural basis of N6-adenosine
methylation by the METTL3-METTL14 complex. _Nature_ 534, 575–578 (2016). Article CAS Google Scholar * Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein
domains. _Nat. Biotechnol._ 33, 661–667 (2015). Article CAS Google Scholar * Wang, T., et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with
oncogenic Ras. _Cell_ 168, 890–903 (2017). Article CAS Google Scholar * Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. _Nat. Methods_
12, 767–772 (2015). Article CAS Google Scholar * Sommer, S., Lavi, U. & Darnell, J.E. Jr . The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases
with label time. _J. Mol. Biol._ 124, 487–499 (1978). Article CAS Google Scholar * Ingolia, N.T., Lareau, L.F. & Weissman, J.S. Ribosome profiling of mouse embryonic stem cells
reveals the complexity and dynamics of mammalian proteomes. _Cell_ 147, 789–802 (2011). Article CAS Google Scholar * Wang, X. et al. N6-methyladenosine modulates messenger RNA translation
efficiency. _Cell_ 161, 1388–1399 (2015). Article CAS Google Scholar * Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide
expression profiles. _Proc. Natl. Acad. Sci. USA_ 102, 15545–15550 (2005). Article CAS Google Scholar * Park, S.M. et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling
in HSCs. _J. Exp. Med._ 211, 71–87 (2014). Article CAS Google Scholar * Li, S. et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia.
_Nat. Med._ 22, 792–799 (2016). Article CAS Google Scholar * Sarry, J.E. et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in
NOD/SCID/IL2Rgc-deficient mice. _J. Clin. Invest._ 121, 384–395 (2011). Article CAS Google Scholar * Stoneley, M. et al. c-Myc protein synthesis is initiated from the internal ribosome
entry segment during apoptosis. _Mol. Cell. Biol._ 20, 1162–1169 (2000). Article CAS Google Scholar * Pandey, S. & Wang, E. Cells en route to apoptosis are characterized by the
upregulation of c-fos, c-myc, c-jun, cdc2, and RB phosphorylation, resembling events of early cell-cycle traverse. _J. Cell. Biochem._ 58, 135–150 (1995). Article CAS Google Scholar *
Sherrill, K.W., Byrd, M.P., Van Eden, M.E. & Lloyd, R.E. BCL-2 translation is mediated via internal ribosome entry during cell stress. _J. Biol. Chem._ 279, 29066–29074 (2004). Article
CAS Google Scholar * Ping, X.L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. _Cell Res._ 24, 177–189 (2014). Article CAS Google Scholar
* Kharas, M.G. et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. _Blood_ 115, 1406–1415 (2010). Article CAS Google Scholar * Sykes, S.M.
et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. _Cell_ 146, 697–708 (2011). Article CAS Google Scholar * Cui, Q. et al. m6A RNA methylation
regulates the self-renewal and tumorigenesis of glioblastoma stem cells. _Cell Reports_ 18, 2622–2634 (2017). Article CAS Google Scholar * Zhang, S., et al. m6A demethylase ALKBH5
maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. _Cancer Cell_ 31, 591–606 (2017). Article CAS Google Scholar *
Kwok, C.T., Marshall, A.D., Rasko, J.E. & Wong, J.J. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. _J. Hematol. Oncol._ 10, 39 (2017).
Article Google Scholar * Patil, D.P. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. _Nature_ 537, 369–373 (2016). Article CAS Google Scholar *
Olarerin-George, A.O. & Jaffrey, S.R. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. _Bioinformatics_ 33, 1563–1564
(2017). Article CAS Google Scholar * Bailey, T.L. et al. MEME SUITE: tools for motif discovery and searching. _Nucleic Acids Res._ 37, W202–208 (2009). Article Google Scholar * Kruse,
S. et al. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. _Sci. Rep._ 1, 126 (2011). Article Google Scholar * Reid, D.W., Shenolikar, S.
& Nicchitta, C.V. Simple and inexpensive ribosome profiling analysis of mRNA translation. _Methods_ 91, 69–74 (2015). Article CAS Google Scholar * Dobin, A. et al. STAR: ultrafast
universal RNA-seq aligner. _Bioinformatics_ 29, 15–21 (2013). Article CAS Google Scholar * Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for
RNA-seq data with DESeq2. _Genome Biol._ 15, 550 (2014). Article Google Scholar Download references ACKNOWLEDGEMENTS We thank D. Bachovchin (MSKCC, New York) for the MOLM-13 constitutively
expressing Cas9 cell line, and C. Vakoc (Cold Spring Harbor Laboratory, New York) for the constitutively expressing Cas9-RN2c cell line. We would like to thank the members of the Scaltriti
laboratory (MSKCC, New York) for providing us with PI3K/AKT inhibitors. We thank the Weill Cornell Medicine Epigenomics Core for their assistance with sequencing. M.G.K. was supported by the
US National Institutes of Health National Institute of Diabetes Digestive and Kidney Diseases Career Development Award, NIDDK NIH R01-DK101989-01A1, NCI 1R01CA193842-01, Kimmel Scholar
Award, V-Scholar Award, Geoffrey Beene Award, Leukemia Lymphoma Society Career Development Award and Alex's Lemonade Stand A Award. This work was also supported by a Tri-Institutional
Stem Cell Award (M.G.K. and S.R.J.), R01CA186702 (S.R.J.), T32CA062948 (B.F.P.), Ruth L. Kirschstein National Research Service Award 1F32CA22104-01 (B.F.P.), a Damon Runyon-Sohn Pediatric
Cancer Fellowship Award DRSG10-14 (L.P.V.), and the American-Italian Cancer Foundation (S.Z.). The research was funded in part through the NIH/NCI Cancer Support Core Grant P30 CA08748 MGK.
The RPPA core facility is funded by NCI #CA16672. AUTHOR INFORMATION Author notes * Ly P Vu, Brian F Pickering and Yuanming Cheng: These authors contributed equally to this work. AUTHORS AND
AFFILIATIONS * Molecular Pharmacology Program, Center for Cell Engineering, Center for Stem Cell Biology, Center for Experimental Therapeutics, Center for Hematologic Malignancies, Memorial
Sloan Kettering Cancer Center, New York, New York, USA Ly P Vu, Yuanming Cheng, Diu Nguyen, Gerard Minuesa, Timothy Chou, Arthur Chow & Michael G Kharas * Department of Pharmacology,
Weill Cornell Medicine, Cornell University, New York, New York, USA Brian F Pickering, Sara Zaccara & Samie R Jaffrey * Department of Physiology and Biophysics, Weill Cornell Medicine,
Cornell University, New York, New York, USA Yogesh Saletore, Matthew MacKay & Christopher E Mason * Department of Medicine, Hematologic Oncology Tissue Bank, Memorial Sloan Kettering
Cancer Center, New York, New York, USA Jessica Schulman * Center for Hematologic Malignancies, Memorial Sloan Kettering Cancer Center, New York, New York, USA Christopher Famulare &
Minal Patel * Department of Medicine, Memorial Sloan Kettering Cancer Center, Leukemia Service, New York, New York, USA Virginia M Klimek * Department of Medicine and Department of
Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, Virginia, USA Francine E Garrett-Bakelman * Division of Hematology and Medical Oncology, Department of Medicine
and Department of Pharmacology, Weill Cornell Medicine, Cornell University, New York, New York, USA Ari Melnick * Division of Hematology and Oncology, University of Pennsylvania,
Philadelphia, Pennsylvania, USA Martin Carroll * The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Cornell Medicine, New York, New York,
USA Christopher E Mason * The Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, New York, New York, USA Christopher E Mason Authors * Ly P Vu View author publications
You can also search for this author inPubMed Google Scholar * Brian F Pickering View author publications You can also search for this author inPubMed Google Scholar * Yuanming Cheng View
author publications You can also search for this author inPubMed Google Scholar * Sara Zaccara View author publications You can also search for this author inPubMed Google Scholar * Diu
Nguyen View author publications You can also search for this author inPubMed Google Scholar * Gerard Minuesa View author publications You can also search for this author inPubMed Google
Scholar * Timothy Chou View author publications You can also search for this author inPubMed Google Scholar * Arthur Chow View author publications You can also search for this author
inPubMed Google Scholar * Yogesh Saletore View author publications You can also search for this author inPubMed Google Scholar * Matthew MacKay View author publications You can also search
for this author inPubMed Google Scholar * Jessica Schulman View author publications You can also search for this author inPubMed Google Scholar * Christopher Famulare View author
publications You can also search for this author inPubMed Google Scholar * Minal Patel View author publications You can also search for this author inPubMed Google Scholar * Virginia M
Klimek View author publications You can also search for this author inPubMed Google Scholar * Francine E Garrett-Bakelman View author publications You can also search for this author
inPubMed Google Scholar * Ari Melnick View author publications You can also search for this author inPubMed Google Scholar * Martin Carroll View author publications You can also search for
this author inPubMed Google Scholar * Christopher E Mason View author publications You can also search for this author inPubMed Google Scholar * Samie R Jaffrey View author publications You
can also search for this author inPubMed Google Scholar * Michael G Kharas View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS L.P.V. led the
project, performed experiments, analyzed data and wrote the manuscript. B.F.P., Y.C., D.N. and S.Z. performed experiments, analyzed data and edited the manuscript. G.M., T.C. and A.C.
provided experimental supports. C.E.M., Y.S. and M.M. performed and analyzed MeRIP-seq experiments on patient-derived samples. J.S., C.F., M.P., F.E.G.-B., A.M., V.M.K. and M.C. provided
patient samples. S.R.J. supervised the project and wrote the manuscript. M.G.K. directed the project, analyzed data and wrote the manuscript. Publisher's note: Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. CORRESPONDING AUTHORS Correspondence to Samie R Jaffrey or Michael G Kharas. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figures 1–6 (PDF 1925 kb) LIFE SCIENCES
REPORTING SUMMARY (PDF 86 KB) SUPPLEMENTARY DATASET Supplementary Tables 1–13 (XLSX 9967 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Vu, L.,
Pickering, B., Cheng, Y. _et al._ The _N_6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. _Nat Med_ 23, 1369–1376
(2017). https://doi.org/10.1038/nm.4416 Download citation * Received: 07 May 2017 * Accepted: 06 September 2017 * Published: 18 September 2017 * Issue Date: 01 November 2017 * DOI:
https://doi.org/10.1038/nm.4416 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative

:max_bytes(150000):strip_icc():focal(584x439:586x441)/amanda-calo-553b00acd15946829c461184e9241390.jpg)



:max_bytes(150000):strip_icc():focal(319x0:321x2)/people_social_image-60e0c8af9eb14624a5b55f2c29dbe25b.png)

