- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
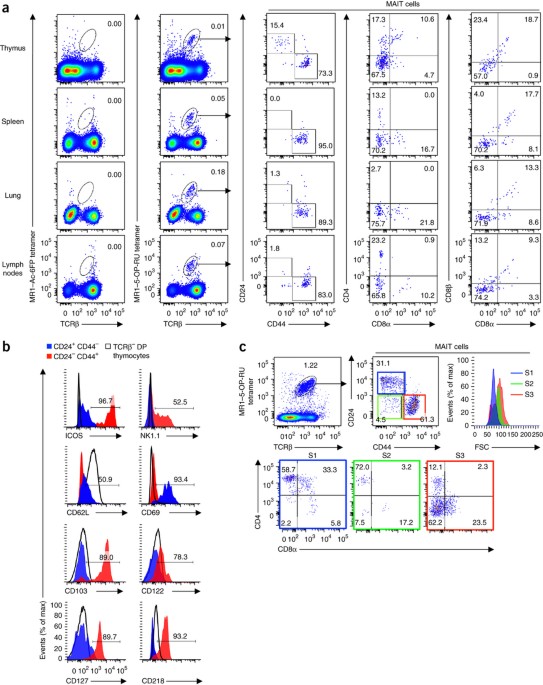
ABSTRACT Mucosal-associated invariant T cells (MAIT cells) detect microbial vitamin B2 derivatives presented by the antigen-presenting molecule MR1. Here we defined three developmental
stages and checkpoints for the MAIT cell lineage in humans and mice. Stage 1 and stage 2 MAIT cells predominated in thymus, while stage 3 cells progressively increased in abundance
extrathymically. Transition through each checkpoint was regulated by MR1, whereas the final checkpoint that generated mature functional MAIT cells was controlled by multiple factors,
including the transcription factor PLZF and microbial colonization. Furthermore, stage 3 MAIT cell populations were expanded in mice deficient in the antigen-presenting molecule CD1d,
suggestive of a niche shared by MAIT cells and natural killer T cells (NKT cells). Accordingly, this study maps the developmental pathway and checkpoints that control the generation of
functional MAIT cells. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution
Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full
article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs *
Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS DIFFERENTIAL CONTROLS OF MAIT CELL EFFECTOR POLARIZATION BY MTORC1/MTORC2 VIA INTEGRATING CYTOKINE AND COSTIMULATORY SIGNALS
Article Open access 01 April 2021 DIFFERENTIAL LOCATION OF NKT AND MAIT CELLS WITHIN LYMPHOID TISSUE Article Open access 08 March 2022 MR1 ANTIGEN PRESENTATION TO MAIT CELLS AND OTHER
MR1-RESTRICTED T CELLS Article 29 September 2023 REFERENCES * Treiner, E. et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. _Nature_ 422, 164–169
(2003). Article CAS Google Scholar * Corbett, A.J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. _Nature_ 509, 361–365 (2014). Article CAS
Google Scholar * Kjer-Nielsen, L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. _Nature_ 491, 717–723 (2012). Article CAS Google Scholar * Tilloy, F. et al. An
invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. _J. Exp. Med._ 189, 1907–1921
(1999). Article CAS Google Scholar * Gherardin, N.A. et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition.
_Immunity_ 44, 32–45 (2016). Article CAS Google Scholar * Tsukamoto, K., Deakin, J.E., Graves, J.A. & Hashimoto, K. Exceptionally high conservation of the MHC class I-related gene,
MR1, among mammals. _Immunogenetics_ 65, 115–124 (2013). Article CAS Google Scholar * Gold, M.C. & Lewinsohn, D.M. Co-dependents: MR1-restricted MAIT cells and their antimicrobial
function. _Nat. Rev. Microbiol._ 11, 14–19 (2013). Article CAS Google Scholar * Dusseaux, M. et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T
cells. _Blood_ 117, 1250–1259 (2011). Article CAS Google Scholar * Le Bourhis, L. et al. Antimicrobial activity of mucosal-associated invariant T cells. _Nat. Immunol._ 11, 701–708
(2010). Article CAS Google Scholar * Tang, X.Z. et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. _J. Immunol._ 190, 3142–3152 (2013).
Article CAS Google Scholar * Martin, E. et al. Stepwise development of MAIT cells in mouse and human. _PLoS Biol._ 7, e54 (2009). Article Google Scholar * Gold, M.C. et al. Human
mucosal associated invariant T cells detect bacterially infected cells. _PLoS Biol._ 8, e1000407 (2010). Article Google Scholar * Meierovics, A., Yankelevich, W.J. & Cowley, S.C. MAIT
cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. _Proc. Natl. Acad. Sci. USA_ 110, E3119–E3128 (2013). Article CAS Google Scholar *
Godfrey, D.I., Uldrich, A.P., McCluskey, J., Rossjohn, J. & Moody, D.B. The burgeoning family of unconventional T cells. _Nat. Immunol._ 16, 1114–1123 (2015). Article CAS Google
Scholar * Seach, N. et al. Double-positive thymocytes select mucosal-associated invariant T cells. _J. Immunol._ 191, 6002–6009 (2013). Article CAS Google Scholar * Rahimpour, A. et al.
Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. _J. Exp. Med._ 212, 1095–1108 (2015). Article CAS Google
Scholar * Ussher, J.E. et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. _Eur. J. Immunol._ 44, 195–203
(2014). Article CAS Google Scholar * Walker, L.J. et al. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. _Blood_ 119, 422–433 (2012). Article CAS
Google Scholar * Leeansyah, E., Loh, L., Nixon, D.F. & Sandberg, J.K. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. _Nat.
Commun._ 5, 3143 (2014). Article Google Scholar * Eckle, S.B. et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. _J. Exp.
Med._ 211, 1585–1600 (2014). Article CAS Google Scholar * Reantragoon, R. et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
_J. Exp. Med._ 210, 2305–2320 (2013). Article CAS Google Scholar * Eckle, S.B. et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant t cells. _J.
Biol. Chem._ 290, 30204–30211 (2015). Article CAS Google Scholar * Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT
cell development. _J. Exp. Med._ 202, 485–492 (2005). Article CAS Google Scholar * Schmitt, T.M. & Zúñiga-Pflücker, J.C. Induction of T cell development from hematopoietic progenitor
cells by delta-like-1 in vitro. _Immunity_ 17, 749–756 (2002). Article CAS Google Scholar * Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the
development of invariant natural killer T cell effector functions. _Nat. Immunol._ 9, 1055–1064 (2008). Article CAS Google Scholar * Kreslavsky, T. et al. TCR-inducible PLZF transcription
factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. _Proc. Natl. Acad. Sci. USA_ 106, 12453–12458 (2009). Article CAS Google Scholar * Savage,
A.K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. _Immunity_ 29, 391–403 (2008). Article CAS Google Scholar * Henao-Mejia, J. et al. The
microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. _Immunity_ 38, 984–997 (2013). Article CAS Google
Scholar * Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. _Nature_ 425, 415–419 (2003). Article CAS Google Scholar * Chong, M.M., Rasmussen, J.P., Rudensky,
A.Y. & Littman, D.R. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. _J. Exp. Med._ 205, 2005–2017 (2008). Article CAS Google Scholar *
Zhou, L. et al. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. _Proc. Natl. Acad. Sci. USA_ 106, 10266–10271 (2009). Article CAS
Google Scholar * Levy, M. et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. _Cell_ 163, 1428–1443 (2015). Article
CAS Google Scholar * Wang, J. et al. Interleukin 18 function in atherosclerosis is mediated by the interleukin 18 receptor and the Na-Cl co-transporter. _Nat. Med._ 21, 820–826 (2015).
Article CAS Google Scholar * Nold-Petry, C.A. et al. IL-37 requires the receptors IL-18Rα and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal
transduction. _Nat. Immunol._ 16, 354–365 (2015). Article CAS Google Scholar * Kawachi, I., Maldonado, J., Strader, C. & Gilfillan, S. MR1-restricted Vα19i mucosal-associated
invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. _J. Immunol._ 176, 1618–1627 (2006). Article CAS Google Scholar * Cui,
Y. et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. _J. Clin. Invest._ 125, 4171–4185 (2015). Article Google Scholar * Engel, I. et al.
Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. _Nat. Immunol._ 17, 728–739 (2016). Article CAS Google Scholar * Lee, Y.J., Holzapfel,
K.L., Zhu, J., Jameson, S.C. & Hogquist, K.A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. _Nat. Immunol._
14, 1146–1154 (2013). CAS Google Scholar * Godfrey, D.I., Stankovic, S. & Baxter, A.G. Raising the NKT cell family. _Nat. Immunol._ 11, 197–206 (2010). Article CAS Google Scholar *
Borg, Z.D. et al. Polymorphisms in the CD1d promoter that regulate CD1d gene expression are associated with impaired NKT cell development. _J. Immunol._ 192, 189–199 (2014). Article CAS
Google Scholar * Gibbs, A. et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. _Mucosal Immunol._
http://dx.doi.org/10.1038/mi.2016.30 (6 April 2016). * Chen, Z. et al. Mucosal-associated invariant T-cell activation and accumulation after _in vivo_ infection depends on microbial
riboflavin synthesis and co-stimulatory signals. _Mucosal Immunol._ http://dx.doi.org/10.1038/mi.2016.39 (4 May 2016). * Dash, P. et al. Paired analysis of TCRα and TCRβ chains at the
single-cell level in mice. _J. Clin. Invest._ 121, 288–295 (2011). Article CAS Google Scholar * Chua, W.J. et al. Endogenous MHC-related protein 1 is transiently expressed on the plasma
membrane in a conformation that activates mucosal-associated invariant T cells. _J. Immunol._ 186, 4744–4750 (2011). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We
thank the staff of the University of Melbourne DMI and MBC flow cytometry facilities and M. Camilleri, D. Taylor and animal house staff for animal husbandry and assistance with genotyping;
J.C. Zúñiga-Pflücker (University of Toronto) for OP9 and OP9-DL1 cells; T. Hansen (Washington University School of Medicine) for the MR1-blocking antibody 8F2.F9; and the clinical research
midwives G. Christophers, G. Pell and R. Murdoch and the Obstetrics and Midwifery staffs of the Mercy Hospital for Women for assistance with collection of the cord blood samples. Supported
by the National Health and Medical Research Council of Australia (1083942, 1013667 and 1016629; CDF 1035858 to A.E.; ECF 1054431 to D.G.P.; Senior Principal Research Fellowships 1020770 and
1027369 to D.I.G. and D.P.F.; Australia Fellowship AF50 to J.R.; CDF2 Fellowship 1047025 to M.C.; CDF2 Fellowship 1023294 to K.K.; and CDF1 Fellowship 1106004 to L.K.M.), the Australian
Research Council (CE140100011 and LE110100106; Future Fellowship FT140100278 to A.P.U.), the Leukaemia Foundation of Australia (Postgraduate Scholarship for N.A.G.), the National Heart
Foundation of Australia (Future Leader Fellowship for C.A.N.-P.), the Hudson Institute (Star Recruitment Fellowship for M.F.N.) and the Ritchie Centre (Victor Yu Fellowship for M.F.N.).
AUTHOR INFORMATION Author notes * Dale I Godfrey and Daniel G Pellicci: These authors jointly directed this work. AUTHORS AND AFFILIATIONS * Department of Microbiology and Immunology, Peter
Doherty Institute for Infection and Immunity, University of Melbourne, Melbourne, Australia Hui-Fern Koay, Nicholas A Gherardin, Liyen Loh, Laura K Mackay, Catarina F Almeida, Brendan E
Russ, Sammy Bedoui, Zhenjun Chen, Alexandra J Corbett, Sidonia B G Eckle, Bronwyn Meehan, Katherine Kedzierska, Stuart P Berzins, James McCluskey, Adam P Uldrich, Dale I Godfrey & Daniel
G Pellicci * Cancer Immunology Research Program, Peter MacCallum Cancer Centre, East Melbourne, Australia Nicholas A Gherardin * Department of Immunology, John Curtin School of Medical
Research, Canberra, Australia Anselm Enders & Chris C Goodnow * Australian Research Council Centre of Excellence for Advanced Molecular Imaging, University of Melbourne, Melbourne,
Australia Laura K Mackay, Adam P Uldrich, Dale I Godfrey & Daniel G Pellicci * Department of Paediatrics, Monash University, Clayton, Australia Claudia A Nold-Petry & Marcel F Nold *
The Ritchie Centre, Hudson Institute of Medical Research, Clayton, Australia Claudia A Nold-Petry & Marcel F Nold * Royal Children's Hospital, Flemington Road, Parkville, Australia
Yves d'Udekem & Igor E Konstantinov * Department of Obstetrics and Gynaecology, Obstetrics, Nutrition and Endocrinology Group, University of Melbourne, Heidelberg, Australia Martha
Lappas * Mercy Perinatal Research Centre, Mercy Hospital for Women, Heidelberg, Australia Martha Lappas * Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia
Ligong Liu & David P Fairlie * Australian Research Council Centre of Excellence for Advanced Molecular Imaging, University of Queensland, Brisbane, Australia Ligong Liu & David P
Fairlie * Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Australia Jamie Rossjohn * Institute of Infection and Immunity, Cardiff
University, School of Medicine, Heath Park, Cardiff, UK Jamie Rossjohn * Australian Research Council Centre of Excellence for Advanced Molecular Imaging, Monash University, Clayton,
Australia Jamie Rossjohn * St Vincent's Institute of Medical Research, Fitzroy, Victoria, Australia, Mark M Chong * Department of Medicine (St Vincent's), University of Melbourne,
Fitzroy, Australia., Mark M Chong * Collaborative Research Network, Federation University, Ballarat, Australia Stuart P Berzins * Fiona Elsey Cancer Research Institute, Ballarat, Australia
Stuart P Berzins * The Division of Molecular Immunology, Walter and Eliza Hall Institute of Medical Research, Parkville, Australia Gabrielle T Belz * Department of Medical Biology,
University of Melbourne, Parkville, Australia Gabrielle T Belz Authors * Hui-Fern Koay View author publications You can also search for this author inPubMed Google Scholar * Nicholas A
Gherardin View author publications You can also search for this author inPubMed Google Scholar * Anselm Enders View author publications You can also search for this author inPubMed Google
Scholar * Liyen Loh View author publications You can also search for this author inPubMed Google Scholar * Laura K Mackay View author publications You can also search for this author
inPubMed Google Scholar * Catarina F Almeida View author publications You can also search for this author inPubMed Google Scholar * Brendan E Russ View author publications You can also
search for this author inPubMed Google Scholar * Claudia A Nold-Petry View author publications You can also search for this author inPubMed Google Scholar * Marcel F Nold View author
publications You can also search for this author inPubMed Google Scholar * Sammy Bedoui View author publications You can also search for this author inPubMed Google Scholar * Zhenjun Chen
View author publications You can also search for this author inPubMed Google Scholar * Alexandra J Corbett View author publications You can also search for this author inPubMed Google
Scholar * Sidonia B G Eckle View author publications You can also search for this author inPubMed Google Scholar * Bronwyn Meehan View author publications You can also search for this author
inPubMed Google Scholar * Yves d'Udekem View author publications You can also search for this author inPubMed Google Scholar * Igor E Konstantinov View author publications You can also
search for this author inPubMed Google Scholar * Martha Lappas View author publications You can also search for this author inPubMed Google Scholar * Ligong Liu View author publications You
can also search for this author inPubMed Google Scholar * Chris C Goodnow View author publications You can also search for this author inPubMed Google Scholar * David P Fairlie View author
publications You can also search for this author inPubMed Google Scholar * Jamie Rossjohn View author publications You can also search for this author inPubMed Google Scholar * Mark M Chong
View author publications You can also search for this author inPubMed Google Scholar * Katherine Kedzierska View author publications You can also search for this author inPubMed Google
Scholar * Stuart P Berzins View author publications You can also search for this author inPubMed Google Scholar * Gabrielle T Belz View author publications You can also search for this
author inPubMed Google Scholar * James McCluskey View author publications You can also search for this author inPubMed Google Scholar * Adam P Uldrich View author publications You can also
search for this author inPubMed Google Scholar * Dale I Godfrey View author publications You can also search for this author inPubMed Google Scholar * Daniel G Pellicci View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS H.-F.K., N.A.G., C.F.A. and D.G.P. performed experiments; H.-F.K. prepared figures; A.E., L.Lo.,
L.K.M., B.E.R., C.A.N.-P., M.F.N., S.B., Z.C., A.J.C., S.B.G.E., B.M., Y.d.U., I.E.K., M.L., L.Li., C.C.G., D.P.F., J.R., M.M.C., K.K., S.P.B., G.T.B. and J.M. facilitated experiments and/or
provided reagents and tissue samples; H.-F.K., A.P.U., D.I.G. and D.G.P. planned experiments, interpreted data and prepared the manuscript; and D.I.G. and D.G.P. led the investigation.
CORRESPONDING AUTHORS Correspondence to Dale I Godfrey or Daniel G Pellicci. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. INTEGRATED
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 RORΓT EXPRESSION ON MAIT CELLS IN RORΓT-GFP REPORTER MICE. Flow cytometric analysis of MAIT cells from MR1-5-OP-RU tetramer enriched
RORγt-GFP reporter mouse thymi for CD24, CD44, and GFP expression. Data are representative of a total of 9 mice with 3-pooled mouse thymi from 2 independent experiments. SUPPLEMENTARY FIGURE
2 THYMIC MAIT CELL SUBSETS IN MICE WITH TRANSGENIC EXPRESSION OF THE TRAV1-TRAJ33 TCR. Flow cytometric analysis of MR1-5-OP-RU tetramer reactive MAIT cells in TRAV1-TRAJ33 Cα-/- TCR
transgenic mouse thymus for expression of CD4, CD8, CD24 and CD44. Data are representative of 3 independent experiments. SUPPLEMENTARY FIGURE 3 NKT CELLS IN DROSHA-DEFICIENT MICE AND
GERM-FREE MICE. (a) Flow cytometric analysis of CD1d-PBS44 tetramer+ TCRβ+ NKT cells from thymus, spleen, lymph nodes from _Drosha__fl/+__ CD4-Cre_ heterozygous control mice and
_Drosha__fl/fl__ CD4-Cre_ mice. Absolute numbers and percentage NKT cells of TCRβ+ cells in thymus, spleen and lymph nodes of _Drosha__fl/+__ CD4-Cre_ heterozygous control mice and
_Drosha__fl/fl__ CD4-Cre_ mice. (b) Flow cytometric analysis of NKT cells from thymus, spleen, lymph nodes from control (SPF) mice and germ-free (GF) mice. Absolute numbers and percentage
NKT cells of TCRβ+ cells in thymus and spleen of SPF mice and GF mice. *P<0.1 **P<0.01 ***P<0.001 NS = not significant (Mann-Whitney rank sum U test (a, b)). Data are representative
of 3 independent experiments with a total of 8 mice per group (a; mean ± SEM) or of 2 independent experiments with a combined total of 11-15 mice per group (b; mean ± SEM). SUPPLEMENTARY
FIGURE 4 MAIT CELLS IN IL-18-DEFICIENT MICE AND IL-18RΑ-DEFICIENT MICE. (a) Flow cytometric analysis of MAIT cells from MR1-5-OP-RU tetramer enriched thymus, spleen and lymph nodes from WT
and IL-18-deficient mice for CD24 and CD44 expression. (b) Absolute numbers and percentage MAIT cells of TCRβ+ cells in individual thymus, spleen and lymph nodes of WT and IL-18-deficient
mice. (c) Flow cytometric analysis of MAIT cells from MR1-5-OP-RU tetramer enriched thymus, spleen and lymph nodes from WT and IL-18Rα-deficient mice for CD24 and CD44 expression. (d)
Absolute numbers and percentage MAIT cells of TCRβ+ cells in individual thymus, spleen and lymph nodes of WT and IL-18Rα-deficient mice. *P<0.1 **P<0.01 ***P<0.001 NS = not
significant (Mann-Whitney rank sum U test (b, d)). Data are representative of 3 independent experiments with a total of 12 mice per group (a, b; mean ± SEM) or 2 independent experiments with
a total of 10 mice per group (c, d; mean ± SEM). SUPPLEMENTARY FIGURE 5 MAIT CELLS IN C57BL/6 CD1D-DEFICIENT MICE. Flow cytometric analysis of MAIT cells from thymus, MR1-5-OP-RU enriched
thymus and spleen from C57BL/6 WT and C57BL/6 CD1d-deficient mice for CD24, CD44 and CD4/CD8 co-receptor expression. Data are representative of 3 independent experiments with a total of 6
mice per group. SUPPLEMENTARY FIGURE 6 SCHEMATIC OF THE THREE DEVELOPMENT STAGES OF MAIT CELLS. Mouse and human MAIT cell development in the thymus can be defined by three separate stages.
Mouse thymic MAIT cells can be defined by a three-stage sequential pathway from CD24+CD44− (stage 1), via CD24−CD44− (stage 2), to CD24−CD44+ (stage 3), while in humans thymic MAIT cells can
be defined by three distinct stages from CD161−CD27− (stage 1), via CD161−CD27+ (stage 2), to CD161+CD27+/lo (stage 3). These stages are regulated by several factors including MR1, PLZF,
Drosha and commensal bacteria. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–6 and Supplementary Tables 1 and 2 (PDF 1562 kb) RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Koay, HF., Gherardin, N., Enders, A. _et al._ A three-stage intrathymic development pathway for the mucosal-associated invariant
T cell lineage. _Nat Immunol_ 17, 1300–1311 (2016). https://doi.org/10.1038/ni.3565 Download citation * Received: 27 May 2016 * Accepted: 22 August 2016 * Published: 26 September 2016 *
Issue Date: November 2016 * DOI: https://doi.org/10.1038/ni.3565 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative