- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
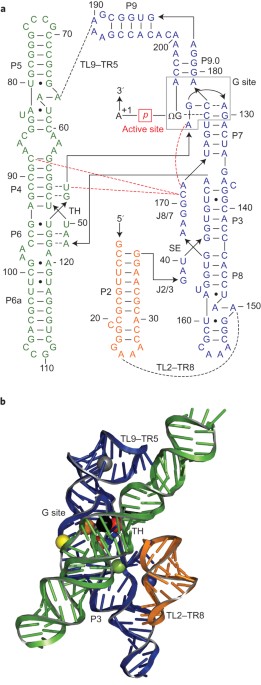
ABSTRACT Ribozymes, which carry out phosphoryl-transfer reactions, often require Mg2+ ions for catalytic activity. The correct folding of the active site and ribozyme tertiary structure is
also regulated by metal ions in a manner that is not fully understood. Here we employ coarse-grained molecular simulations to show that individual structural elements of the group I ribozyme
from the bacterium _Azoarcus_ form spontaneously in the unfolded ribozyme even at very low Mg2+ concentrations, and are transiently stabilized by the coordination of Mg2+ ions to specific
nucleotides. However, competition for scarce Mg2+ and topological constraints that arise from chain connectivity prevent the complete folding of the ribozyme. A much higher Mg2+
concentration is required for complete folding of the ribozyme and stabilization of the active site. When Mg2+ is replaced by Ca2+ the ribozyme folds, but the active site remains unstable.
Our results suggest that group I ribozymes utilize the same interactions with specific metal ligands for both structural stability and chemical activity. Access through your institution Buy
or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and
online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes
which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY
OTHERS MAGNESIUM IONS MEDIATE LIGAND BINDING AND CONFORMATIONAL TRANSITION OF THE SAM/SAH RIBOSWITCH Article Open access 31 July 2023 A HAMMERHEAD RIBOZYME SELECTS MECHANICALLY STABLE
CONFORMATIONS FOR CATALYSIS AGAINST VIRAL RNA Article Open access 03 February 2025 CATION-INDUCED INTRAMOLECULAR COIL-TO-GLOBULE TRANSITION IN POLY(ADP-RIBOSE) Article Open access 10
September 2024 REFERENCES * Cech, T. R., Zaug, A. J. & Grabowski, P. J. _In vitro_ splicing of the ribosomal-RNA precursor of _Tetrahymena_—involvement of a guanosine nucleotide in the
excision of the intervening sequence. _Cell_ 27, 487–496 (1981). Article CAS Google Scholar * Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. The RNA moiety of
ribonuclease-P is the catalytic subunit of the enzyme. _Cell_ 35, 849–857 (1983). Article CAS Google Scholar * Doudna, J. & Cech, T. The chemical repertoire of natural ribozymes.
_Nature_ 418, 222–228 (2002). Article CAS Google Scholar * Piccirilli, J. A., Vyle, J. S., Caruthers, M. H. & Cech, T. R. Metal ion catalysis in the _Tetrahymena_ ribozyme reaction.
_Nature_ 361, 85–88 (1993). Article CAS Google Scholar * Weinstein, L. B., Jones, B. C. N. M., Cosstick, R. & Cech, T. R. A second catalytic metal ion in a group I ribozyme. _Nature_
388, 805–808 (1997). Article CAS Google Scholar * Shan, S., Yoshida, A., Sun, S., Piccirilli, J. A. & Herschlag, D. Three metal ions at the active site of the _Tetrahymena_ group I
ribozyme. _Proc. Natl Acad. Sci. USA_ 96, 12299–12304 (1999). Article CAS Google Scholar * Stahley, M. R. & Strobel, S. A. Structural evidence for a two-metal–ion mechanism of group I
intron splicing. _Science_ 309, 1587–1590 (2005). Article CAS Google Scholar * Wilcox, J. L., Ahluwalia, A. K. & Bevilacqua, P. C. Charged nucleobases and their potential for RNA
catalysis. _Acc. Chem. Res._ 44, 1270–1279 (2011). Article CAS Google Scholar * Bowman, J. C., Lenz, T. K., Hud, N. V. & Williams, L. D. Cations in charge: magnesium ions in RNA
folding and catalysis. _Curr. Opin. Struct. Biol._ 22, 262–272 (2012). Article CAS Google Scholar * Orr, J. W., Hagerman, P. J. & Williamson, J. R. Protein and Mg2+-induced
conformational changes in the S15 binding site of 16S ribosomal RNA. _J. Mol. Biol._ 275, 453–464 (1998). Article CAS Google Scholar * Treiber, D. K., Rook, M. S., Zarrinkar, P. P. &
Williamson, J. R. Kinetic intermediates trapped by native interactions in RNA folding. _Science_ 279, 1943–1946 (1998). Article CAS Google Scholar * Rangan, P. & Woodson, S. A.
Structural requirement for Mg2+ binding in the group I intron core. _J. Mol. Biol._ 329, 229–238 (2003). Article CAS Google Scholar * Draper, D. E. RNA folding: thermodynamic and
molecular descriptions of the roles of ions. _Biophys. J._ 95, 5489–5495 (2008). Article CAS Google Scholar * Behrouzi, R., Roh, J. H., Kilburn, D., Briber, R. M. & Woodson, S. A.
Cooperative tertiary interaction network guides RNA folding. _Cell_ 149, 348–357 (2012). Article CAS Google Scholar * Heilman-Miller, S. L., Thirumalai, D. & Woodson, S. A. Role of
counterion condensation in folding of the _Tetrahymena_ ribozyme. I. Equilibrium stabilization by cations. _J. Mol. Biol._ 306, 1157–1166 (2001). Article CAS Google Scholar * Fedorova,
O., Waldsich, C. & Pyle, A. M. Group II intron folding under near-physiological conditions: collapsing to the near-native state. _J. Mol. Biol._ 366, 1099–1114 (2007). Article CAS
Google Scholar * Tan, Z.-J. & Chen, S.-J. Salt contribution to RNA tertiary structure folding stability. _Biophys. J._ 101, 176–187 (2011). Article CAS Google Scholar * Chen, S.-J.
RNA folding: conformational statistics, folding kinetics, and ion electrostatics. _Ann. Rev. Biophys._ 37, 197–214 (2008). Article CAS Google Scholar * Kirmizialtin, S., Pabit, S. A.,
Meisburger, S. P., Pollack, L. & Elber, R. RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations. _Biophys. J._ 102, 819–828 (2012). Article CAS
Google Scholar * Butcher, S. E. & Pyle, A. M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. _Acc. Chem. Res._ 44, 1302–1311
(2011). Article CAS Google Scholar * Tanner, M. A. & Cech, T. R. Activity and thermostability of the small self-splicing group I intron in the pre-tRNAIle of the purple bacterium
_Azoarcus_. _RNA_ 2, 74–83 (1996). CAS PubMed PubMed Central Google Scholar * Adams, P. L., Stahley, M. R., Kosek, A. B., Wang, J. & Strobel, S. A. Crystal structure of a
self-splicing group I intron with both exons. _Nature_ 430, 45–50 (2004). Article CAS Google Scholar * Stahley, M. R., Adams, P. L., Wang, J. & Strobel, S. A. Structural metals in the
group I intron: a ribozyme with a multiple metal ion core. _J. Mol. Biol._ 372, 89–102 (2007). Article CAS Google Scholar * Cate, J. H. et al. Crystal structure of a group I ribozyme
domain: principles of RNA packing. _Science_ 273, 1678–1685 (1996). Article CAS Google Scholar * Guo, F., Gooding, A. R. & Cech, T. R. Structure of the _Tetrahymena_ ribozyme: base
triple sandwich and metal ion at the active site. _Mol. Cell_ 16, 351–362 (2004). CAS PubMed Google Scholar * Takamoto, K., He, Q., Morris, S., Chance, M. R. & Brenowitz, M.
Monovalent cations mediate formation of native tertiary structure of the _Tetrahymena thermophila_ ribozyme. _Nature Struct. Biol._ 9, 928–933 (2002). Article CAS Google Scholar *
Chauhan, S., Behrouzi, R., Rangan, P. & Woodson, S. A. Structural rearrangements linked to global folding pathways of the _Azoarcus_ group I ribozyme. _J. Mol. Biol._ 386, 1167–1178
(2009). Article CAS Google Scholar * Shaw, D. et al. Atomic-level characterization of the structural dynamics of proteins. _Science_ 330, 341–346 (2010). Article CAS Google Scholar *
Lindorff-Larsen, K., Piana, S., Dror, R. O. & Shaw, D. E. How fast-folding proteins fold. _Science_ 334, 517–520 (2011). Article CAS Google Scholar * Lane, T. J., Shukla, D.,
Beauchamp, K. A. & Pande, V. S. To milliseconds and beyond: challenges in the simulation of protein folding. _Curr. Opin. Struct. Biol._ 23, 58–65 (2012). Article Google Scholar *
Chen, A. A. & Garcia, A. E. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. _Proc. Natl Acad. Sci. USA_ 110, 16820–16825 (2013).
Article CAS Google Scholar * Hyeon, C. & Thirumalai, D. Mechanical unfolding of RNA hairpins. _Proc. Natl Acad. Sci. USA_ 102, 6789–6794 (2005). Article CAS Google Scholar * Cao,
S. & Chen, S.-J. Predicting RNA folding thermodynamics with a reduced chain representation model. _RNA_ 11, 1884–1897 (2005). Article CAS Google Scholar * Denesyuk, N. A. &
Thirumalai, D. Coarse-grained model for predicting RNA folding thermodynamics. _J. Phys. Chem. B_ 117, 4901–4911 (2013). Article CAS Google Scholar * Gō, N. & Abe, H. Noninteracting
local-structure model of folding and unfolding transition in globular proteins. I. Formulation. _Biopolymers_ 20, 991–1011 (1981). Article Google Scholar * Whitford, P. C. et al. Nonlocal
helix formation is key to understanding _S_-adenosylmethionine-1 riboswitch function. _Biophys. J._ 96, L7–L9 (2009). Article Google Scholar * Feng, J., Walter, N. G. & Brooks, C. L.
III . Cooperative and directional folding of the preQ(1) riboswitch aptamer domain. _J. Am. Chem. Soc._ 133, 4196–4199 (2011). Article CAS Google Scholar * Lin, J.-C. & Thirumalai, D.
Relative stability of helices determines the folding landscape of adenine riboswitch aptamers. _J. Am. Chem. Soc._ 130, 14080–14081 (2008). Article CAS Google Scholar * Cho, S. S.,
Pincus, D. L. & Thirumalai, D. Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. _Proc. Natl Acad. Sci. USA_ 106, 17349–17354
(2009). Article CAS Google Scholar * Whitford, P. C. et al. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. _RNA_ 16, 1196–1204
(2010). Article CAS Google Scholar * Denesyuk, N. A. & Thirumalai, D. Crowding promotes the switch from hairpin to pseudoknot conformation in human telomerase RNA. _J. Am. Chem. Soc._
133, 11858–11861 (2011). Article CAS Google Scholar * Hayes, R. L. et al. Reduced model captures Mg2+–RNA interaction free energy of riboswitches. _Biophys. J._ 106, 1508–1519 (2014).
Article CAS Google Scholar * Karbstein, K. & Herschlag, D. Extraordinarily slow binding of guanosine to the _Tetrahymena_ group I ribozyme: implications for RNA preorganization and
function. _Proc. Natl Acad. Sci. USA_ 100, 2300–2305 (2003). Article CAS Google Scholar * Rangan, P., Masquida, B., Westhof, E. & Woodson, S. A. Assembly of core helices and rapid
tertiary folding of a small bacterial group I ribozyme. _Proc. Natl Acad. Sci. USA_ 100, 1574–1579 (2003). Article CAS Google Scholar * Chauhan, S. et al. RNA tertiary interactions
mediate native collapse of a bacterial group I ribozyme. _J. Mol. Biol._ 353, 1199–1209 (2005). Article CAS Google Scholar * Jaeger, L., Westhof, E. & Michel, F. Monitoring of the
cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3′ terminal intron components. _J. Mol.
Biol._ 234, 331–346 (1993). Article CAS Google Scholar * Hasted, J. B. in _Water, a Comprehensive Treatise_ Vol. 1 (ed. Franks, F.) 255–309 (Plenum Press, 1972). Google Scholar * Roh,
J. H. et al. Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering. _J. Am. Chem. Soc._ 132, 10148–10154 (2010). Article CAS Google Scholar
Download references ACKNOWLEDGEMENTS This work was supported by a grant from the National Science Foundation (CHE 13-61946). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute for
Physical Science and Technology, University of Maryland, College Park, 20742, Maryland, USA Natalia A. Denesyuk & D. Thirumalai * Department of Chemistry and Biochemistry and Biophysics
Program, University of Maryland, College Park, 20742, Maryland, USA D. Thirumalai Authors * Natalia A. Denesyuk View author publications You can also search for this author inPubMed Google
Scholar * D. Thirumalai View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS N.A.D. and D.T. conceived and designed the project, analysed the
simulation data and co-wrote the paper. N.A.D. performed the simulations. CORRESPONDING AUTHOR Correspondence to D. Thirumalai. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary information (PDF 834 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Denesyuk, N., Thirumalai, D. How do metal ions direct ribozyme folding?. _Nature Chem_ 7, 793–801 (2015). https://doi.org/10.1038/nchem.2330 Download citation * Received:
11 September 2014 * Accepted: 20 July 2015 * Published: 31 August 2015 * Issue Date: October 2015 * DOI: https://doi.org/10.1038/nchem.2330 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative