- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
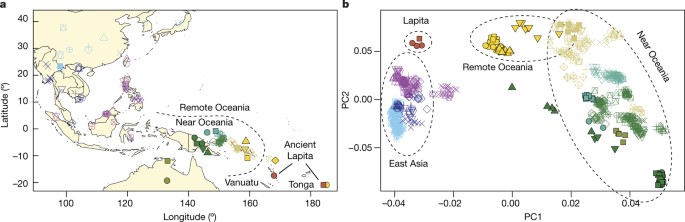
ABSTRACT The appearance of people associated with the Lapita culture in the South Pacific around 3,000 years ago1 marked the beginning of the last major human dispersal to unpopulated lands.
However, the relationship of these pioneers to the long-established Papuan people of the New Guinea region is unclear. Here we present genome-wide ancient DNA data from three individuals
from Vanuatu (about 3,100–2,700 years before present) and one from Tonga (about 2,700–2,300 years before present), and analyse them with data from 778 present-day East Asians and Oceanians.
Today, indigenous people of the South Pacific harbour a mixture of ancestry from Papuans and a population of East Asian origin that no longer exists in unmixed form, but is a match to the
ancient individuals. Most analyses have interpreted the minimum of twenty-five per cent Papuan ancestry in the region today as evidence that the first humans to reach Remote Oceania,
including Polynesia, were derived from population mixtures near New Guinea, before their further expansion into Remote Oceania2,3,4,5. However, our finding that the ancient individuals had
little to no Papuan ancestry implies that later human population movements spread Papuan ancestry through the South Pacific after the first peopling of the islands. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print
issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS MEDIEVAL DNA FROM SOQOTRA POINTS TO EURASIAN ORIGINS OF AN ISOLATED POPULATION AT THE CROSSROADS OF AFRICA AND ARABIA Article 08 February 2024 PALAEOGENOMIC INSIGHTS
INTO THE ORIGINS OF EARLY SETTLERS ON THE ISLAND OF CYPRUS Article Open access 26 April 2024 THE GENOMIC LANDSCAPE OF MEXICAN INDIGENOUS POPULATIONS BRINGS INSIGHTS INTO THE PEOPLING OF THE
AMERICAS Article Open access 12 October 2021 ACCESSION CODES PRIMARY ACCESSIONS EUROPEAN NUCLEOTIDE ARCHIVE * PRJEB14728 DATA DEPOSITS The aligned sequences are available through the
European Nucleotide Archive under accession number PRJEB14728. The newly reported SNP genotyping data for the subset of individuals who provided informed consent consistent with fully public
distribution are available at http://genetics.med.harvard.edu/reichlab/Reich_Lab/Datasets.html. To access data for the remaining samples, researchers should send a signed letter to D.R.
containing the following text: “(a) I will not distribute the data outside my collaboration; (b) I will not post the data publicly; (c) I will make no attempt to connect the genetic data to
personal identifiers for the samples; (d) I will use the data only for studies of population history; (e) I will not use the data for any selection studies; (f) I will not use the data for
medical or disease-related analyses; (g) I will not use the data for commercial purposes.” Extended Data Table 2 specifies which samples are consistent with which type of data distribution.
REFERENCES * Sheppard, P. J., Chiu, S. & Walter, R. Re-dating Lapita movement into Remote _Oceania. J. Pacific Archaeol._ 6, 26–36 (2015) Google Scholar * Kayser, M. et al. Genome-wide
analysis indicates more Asian than Melanesian ancestry of Polynesians. _Am. J. Hum. Genet._ 82, 194–198 (2008) Article CAS PubMed PubMed Central Google Scholar * Kayser, M. The human
genetic history of Oceania: near and remote views of dispersal. _Curr. Biol._ 20, R194–R201 (2010) Article CAS PubMed Google Scholar * Wollstein, A. et al. Demographic history of Oceania
inferred from genome-wide data. _Curr. Biol._ 20, 1983–1992 (2010) Article CAS PubMed Google Scholar * Matisoo-Smith, E. Ancient DNA and the human settlement of the Pacific: a review.
_J. Hum. Evol._ 79, 93–104 (2015) Article PubMed Google Scholar * Bellwood, P. S. _First Farmers: the Origins of Agricultural Societies_ (Blackwell Publishing, 2005) * Duggan, A. T. et
al. Maternal history of Oceania from complete mtDNA genomes: contrasting ancient diversity with recent homogenization due to the Austronesian expansion. _Am. J. Hum. Genet._ 94, 721–733
(2014) Article CAS PubMed PubMed Central Google Scholar * Kayser, M. et al. Melanesian origin of Polynesian Y chromosomes. _Curr. Biol._ 10, 1237–1246 (2000) Article CAS PubMed
Google Scholar * Blust, R. Remote Melanesia: one history or two? An addendum to Donohue and Denham. _Oceanic Linguistics_ 47, 445–459 (2008) Article Google Scholar * Friedlaender, J. S.
et al. The genetic structure of Pacific Islanders. _PLoS Genet._ 4, e19 (2008) Article CAS PubMed PubMed Central Google Scholar * Pinhasi, R. et al. Optimal ancient DNA yields from the
inner ear part of the human petrous bone. _PLoS One_ 10, e0129102 (2015) Article CAS PubMed PubMed Central Google Scholar * Dabney, J. et al. Complete mitochondrial genome sequence of a
Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. _Proc. Natl Acad. Sci. USA_ 110, 15758–15763 (2013) Article ADS PubMed PubMed Central Google Scholar *
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Phil. _Trans. R. Soc. Lond. B_ 370, 20130624
(2015) Article CAS Google Scholar * Fu, Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. _Nature_ 524, 216–219 (2015) Article ADS CAS PubMed PubMed
Central Google Scholar * Skoglund, P., Storå, J., Götherström, A. & Jakobsson, M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. _J. Archaeol. Sci._
40, 4477–4482 (2013) Article CAS Google Scholar * Melton, T. et al. Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis. _Am. J. Hum. Genet._
57, 403–414 (1995) Article CAS PubMed PubMed Central Google Scholar * Skoglund, P. et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. _Science_ 336,
466–469 (2012) Article ADS CAS PubMed Google Scholar * Skoglund, P. et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. _Proc. Natl Acad.
Sci. USA_ 111, 2229–2234 (2014) Article ADS CAS PubMed PubMed Central Google Scholar * Reich, D. et al. Reconstructing Native American population history. _Nature_ 488, 370–374 (2012)
Article ADS CAS PubMed PubMed Central Google Scholar * Moorjani, P. et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. _PLoS Genet._ 7, e1001373
(2011) Article CAS PubMed PubMed Central Google Scholar * Loh, P.-R. et al.Inferring admixture histories of human populations using linkage disequilibrium. _Genetics_ 193, 1233–1254
(2013) Article PubMed PubMed Central Google Scholar * Fenner, J. N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies.
_Am. J. Phys. Anthropol._ 128, 415–423 (2005) Article PubMed Google Scholar * Xu, S., Pugach, I., Stoneking, M., Kayser, M. & Jin, L. Genetic dating indicates that the Asian-Papuan
admixture through Eastern Indonesia corresponds to the Austronesian expansion. _Proc. Natl Acad. Sci. USA_ 109, 4574–4579 (2012) Article ADS PubMed PubMed Central Google Scholar *
Pugach, I., Matveyev, R., Wollstein, A., Kayser, M. & Stoneking, M. Dating the age of admixture via wavelet transform analysis of genome-wide data. _Genome Biol._ 12, R19 (2011) Article
PubMed PubMed Central Google Scholar * Lipson, M. et al. Reconstructing Austronesian population history in Island Southeast Asia. _Nat. Commun._ 5, 4689 (2014) Article ADS CAS PubMed
Google Scholar * Patterson, N. et al. Ancient admixture in human history. _Genetics_ 192, 1065–1093 (2012) Article PubMed PubMed Central Google Scholar * Pickrell, J. K. &
Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. _PLoS Genet._ 8, e1002967 (2012) Article CAS PubMed PubMed Central Google Scholar *
Bellwood, P. Holocene population history in the Pacific region as a model for worldwide food producer dispersals. _Curr. Anthropol._ 52, S363–S378 (2011) Article Google Scholar * Jordan,
F. M., Gray, R. D., Greenhill, S. J. & Mace, R. Matrilocal residence is ancestral in Austronesian societies. _Proc. R. Soc. Lond. B_ 276, 1957–1964 (2009) Article Google Scholar *
Stephen Lansing, J. et al. An ongoing Austronesian expansion in Island Southeast Asia. _J. Anthropol. Archaeol._ 30, 262–272 (2011) Article Google Scholar * Knapp, M., Clarke, A. C.,
Horsburgh, K. A. & Matisoo-Smith, E. A. Setting the stage – Building and working in an ancient DNA laboratory. Ann. Anat. _Anat. Anz._ 194, 3–6 (2012) Article CAS Google Scholar *
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. _Cold Spring Harbor Prot._ http://dx.doi.org/10.1101/pdb.prot5448
(2010) * Fu, Q. et al. DNA analysis of an early modern human from Tianyuan Cave, China. _Proc. Natl Acad. Sci. USA_ 110, 2223–2227 (2013) Article ADS PubMed PubMed Central Google Scholar
* Haak, W. et al. Massive migration from the steppe is a source for Indo-European languages in Europe. _Nature_ 522, 207–211 (2015) Article ADS CAS PubMed PubMed Central Google
Scholar * Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. _Bioinformatics_ 25, 1754–1760 (2009) Article CAS PubMed PubMed Central Google
Scholar * Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. _Nature_ 513, 409–413 (2014) Article ADS CAS PubMed PubMed Central
Google Scholar * Qin, P. & Stoneking, M. Denisovan ancestry in East Eurasian and Native American populations. _Mol. Biol. Evol._ 32, 2665–2674 (2015) Article CAS PubMed Google
Scholar * Skoglund, P. et al. Genetic evidence for two founding populations of the Americas. _Nature_ 525, 104–108 (2015) Article ADS CAS PubMed PubMed Central Google Scholar *
Moreno-Mayar, J. V. et al. Genome-wide ancestry patterns in Rapanui suggest pre-European admixture with Native Americans. _Curr. Biol._ 24, 2518–2525 (2014) Article CAS PubMed Google
Scholar * Reich, D. et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. _Am. J. Hum. Genet._ 89, 516–528 (2011) Article CAS PubMed PubMed
Central Google Scholar * Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. _Nature_ 468, 1053–1060 (2010) Article ADS CAS PubMed PubMed
Central Google Scholar * Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. _Science_ 338, 222–226 (2012) Article ADS CAS PubMed PubMed Central
Google Scholar * Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. _Nature_ 505, 43–49 (2014) Article ADS CAS PubMed Google Scholar * Allentoft,
M. E. et al. Population genomics of Bronze Age Eurasia. _Nature_ 522, 167–172 (2015) Article ADS CAS PubMed Google Scholar * Green, R. E. et al. A draft sequence of the Neandertal
genome. _Science_ 328, 710–722 (2010) Article ADS CAS PubMed PubMed Central Google Scholar * Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native
Americans. _Nature_ 505, 87–91 (2014) Article ADS CAS PubMed Google Scholar * Raghavan, M. et al. The genetic prehistory of the New World Arctic. Science 345, 1255832 (2014) Article
CAS PubMed Google Scholar * Rasmussen, M. et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. _Nature_ 506, 225–229 (2014) Article ADS CAS
PubMed PubMed Central Google Scholar * Rasmussen, M. et al. The ancestry and affiliations of Kennewick Man. _Nature_ 523, 455–458 (2015) Article ADS PubMed PubMed Central Google
Scholar * Skoglund, P. et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. _Science_ 344, 747–750 (2014) Article ADS CAS PubMed Google
Scholar * Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. _PLoS Genet._ 2, e190 (2006) Article CAS PubMed PubMed Central Google Scholar * Skoglund,
P. & Jakobsson, M. Archaic human ancestry in East Asia. _Proc. Natl Acad. Sci. USA_ 45, 18301–18306 (2011) Article ADS Google Scholar * Sudmant, P. H. et al. Global diversity,
population stratification, and selection of human copy-number variation. _Science_ 349, 1174–1181 (2015) Article CAS Google Scholar * Kong, A. et al. A high-resolution recombination map
of the human genome. _Nat. Genet._ 31, 241–247 (2002) Article CAS PubMed Google Scholar * Moorjani, P. et al. A genetic method for dating ancient genomes provides a direct estimate of
human generation interval in the last 45,000 years. _Proc. Natl Acad. Sci. USA_ 113, 5652–5657 (2016) Article ADS CAS PubMed PubMed Central Google Scholar * Reich, D., Thangaraj, K.,
Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. _Nature_ 461, 489–494 (2009) Article ADS CAS PubMed PubMed Central Google Scholar * Csilléry, K.,
Blum, M. G. B., Gaggiotti, O. E. & François, O. Approximate Bayesian Computation (ABC) in practice. _Trends Ecol. Evol._ 25, 410–418 (2010) Article PubMed Google Scholar * Bronk
Ramsey, C. _OxCal Program v4.2.4._ (Radiocarbon Accelerator Unit, Univ. Oxford, 2016) * Reimer, P. J. et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 Years cal BP.
Radiocarbon http://dx.doi.org/10.2458/azu_js_rc.55.16947 (2013) * Ambrose, S. H. Isotopic analysis of paleodiets: methodological and interpretive considerations in _Food and Nutrition in
History and Anthropology_ (USA) http://agris.fao.org/aos/records/US9514697?output=xml (1993) * Petchey, F., Spriggs, M., Bedford, S., Valentin, F. & Buckley, H. Radiocarbon dating of
burials from the Teouma Lapita cemetery, Efate, Vanuatu. _J. Archaeol. Sci._ 50, 227–242 (2014) Article CAS Google Scholar * Petchey, F., Anderson, A., Zondervan, A., Ulm, S. & Hogg,
A. New marine∆ R values for the South Pacific subtropical gyre region. _Radiocarbon_ 50, 373–397 (2008) Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank the 356
volunteers who donated samples for genome-wide analysis; M. Stoneking for co-funding genotyping of the Bismarck samples; M. Brilliant, H. Norton, and L. Scheinfeldt, for help in the
preparation of the Bismarck samples and establishment of a repository for them at the Marshfield Foundation; A. Wissgott for help in data generation from the ancient Tongan individual; A.
Kim, I. Pugach, and M. Stoneking for comments, and I. Mathieson for critiques and advice on estimating sex-specific ancestral contributions. The maps in Figs 1a and 3b–e maps were plotted in
R using the _world()_ map of the ‘fields’ and ‘maps’ packages (using public domain data from the CIA World Data Bank II). P.S. was supported by the Wenner-Gren foundation, SciLifeLab, and
the Swedish Research Council (VR grant 2014-453). The Teouma research by M.S. and S.B. was supported by the Australian Research Council (Discovery Grants DP0880789 and DP110101415), the
National Geographic Society, and the Australia-Pacific Science Foundation. F.V. was supported by CNRS-UMR 7041. M.N. was supported by an Irish Research Council grant (GOIPD/2013/1). D.F. was
supported by an Irish Research Council grant (GOIPG/2013/36). Q.F. was funded by the Key Research Program of Frontier Sciences of CAS (QYZDB-SS W-DQC003), the National Natural Science
Foundation of China (L1524016) and the Chinese Academy of Sciences Discipline Development Strategy Project (2015-DX-C-03). T.K. was supported by ERC starting grant FP7-261213. C.P. and J.K.
were supported by the Baden Wuerttemberg Foundation. J.K was supported by the DFG grant KR 4015/1-1 and the Max Planck Society. R.P. was supported by ERC starting grant ADNABIOARC (263441).
D.R. was supported by NIH grant GM100233, by NSF HOMINID BCS-1032255, and is a Howard Hughes Medical Institute investigator. AUTHOR INFORMATION Author notes * Ron Pinhasi and David Reich:
These authors jointly supervised this work. AUTHORS AND AFFILIATIONS * Department of Genetics, Harvard Medical School, Boston, 02115, Massachusetts, USA Pontus Skoglund, Qiaomei Fu, Eadaoin
Harney, Mark Lipson, Swapan Mallick, Nadin Rohland, Kristin Stewardson & David Reich * Broad Institute of MIT and Harvard, Cambridge, 02142, Massachusetts, USA Pontus Skoglund, Eadaoin
Harney, Swapan Mallick, Kristin Stewardson, Nick Patterson & David Reich * Department of Archaeology and Classical Studies, Archaeological Research Laboratory, Stockholm University,
Stockholm, 10691, Sweden Pontus Skoglund * Institute for Archaeological Sciences, Archaeo- and Palaeogenetics, University of Tübingen, Tübingen, 72070, Germany Cosimo Posth * Max Planck
Institute for the Science of Human History, Jena, 07745, Germany Cosimo Posth & Johannes Krause * School of Archaeology and Earth Institute, Belfield, University College Dublin, Dublin
4, Dublin, Ireland Kendra Sirak, Daniel Fernandes, Mario Novak & Ron Pinhasi * Department of Anthropology, Emory University, Atlanta, 30322, Georgia, USA Kendra Sirak * School of
Archaeology and Anthropology, College of Arts and Social Sciences, The Australian National University, Canberra, 2601, Australian Capital Territory, Australia Matthew Spriggs * Vanuatu
National Museum, Vanuatu Cultural Centre, Port Vila, Vanuatu Matthew Spriggs & Stuart Bedford * Maison de l’Archéologie et de l’Ethnologie, CNRS, UMR 7041, Nanterre, 92023, France
Frederique Valentin * Department of Archaeology and Natural History, College of Asia and the Pacific, The Australian National University, Canberra, 2601, Australian Capital Territory,
Australia Stuart Bedford & Geoffrey R. Clark * College of Arts, Society and Education, James Cook University, Queensland, 4870, Australia Christian Reepmeyer * Radiocarbon Dating
Laboratory, University of Waikato, Hamilton, 3240, New Zealand Fiona Petchey * Department of Life Sciences, CIAS, University of Coimbra, Coimbra, 3000-456, Portugal Daniel Fernandes * Key
Laboratory of Vertebrate Evolution and Human Origins of Chinese Academy of Sciences, IVPP, CAS, Beijing, 100044, China Qiaomei Fu * Department of Evolutionary Genetics, Max Planck Institute
for Evolutionary Anthropology, Leipzig, 04103, Germany Qiaomei Fu * Institute for Anthropological Research, Zagreb, 10000, Croatia Mario Novak * Howard Hughes Medical Institute, Harvard
Medical School, Boston, 02115, Massachusetts, USA Kristin Stewardson & David Reich * RIPAS Hospital, Bandar Seri Begawan, Brunei, Darussalam Syafiq Abdullah * Institute of Fundamental
Sciences, Massey University, Palmerston North, 4442, New Zealand Murray P. Cox * Independent Scientist, Sharon, Connecticut, 06069, USA Françoise R. Friedlaender * Department of
Anthropology, Temple University, Gladfelter Hall, Philadelphia, 19122, Pennsylvania, USA Jonathan S. Friedlaender * Estonian Biocentre, Evolutionary Biology group, Tartu, 51010, Estonia
Toomas Kivisild * Division of Archaeology, University of Cambridge, Fitzwilliam Street, Cambridge, CB2 1QH, UK Toomas Kivisild * Papua New Guinea Institute of Medical Research, Eastern
Highlands Province 441, Goroka, Papua New Guinea George Koki * Eijkman Institute for Molecular Biology, Jakarta, 10430, Indonesia Pradiptajati Kusuma * Department of Anthropology, Binghamton
University, Binghamton, 13902, New York, USA D. Andrew Merriwether * Evolutionary Medicine Group, Laboratoire d’Anthropologie Moléculaire et Imagerie de Synthèse UMR 5288 CNRS, Université
de Toulouse, Toulouse, 31073, France Francois-X. Ricaut * National Cancer Centre Singapore, Singapore, 169610, Singapore Joseph T. S. Wee Authors * Pontus Skoglund View author publications
You can also search for this author inPubMed Google Scholar * Cosimo Posth View author publications You can also search for this author inPubMed Google Scholar * Kendra Sirak View author
publications You can also search for this author inPubMed Google Scholar * Matthew Spriggs View author publications You can also search for this author inPubMed Google Scholar * Frederique
Valentin View author publications You can also search for this author inPubMed Google Scholar * Stuart Bedford View author publications You can also search for this author inPubMed Google
Scholar * Geoffrey R. Clark View author publications You can also search for this author inPubMed Google Scholar * Christian Reepmeyer View author publications You can also search for this
author inPubMed Google Scholar * Fiona Petchey View author publications You can also search for this author inPubMed Google Scholar * Daniel Fernandes View author publications You can also
search for this author inPubMed Google Scholar * Qiaomei Fu View author publications You can also search for this author inPubMed Google Scholar * Eadaoin Harney View author publications You
can also search for this author inPubMed Google Scholar * Mark Lipson View author publications You can also search for this author inPubMed Google Scholar * Swapan Mallick View author
publications You can also search for this author inPubMed Google Scholar * Mario Novak View author publications You can also search for this author inPubMed Google Scholar * Nadin Rohland
View author publications You can also search for this author inPubMed Google Scholar * Kristin Stewardson View author publications You can also search for this author inPubMed Google Scholar
* Syafiq Abdullah View author publications You can also search for this author inPubMed Google Scholar * Murray P. Cox View author publications You can also search for this author inPubMed
Google Scholar * Françoise R. Friedlaender View author publications You can also search for this author inPubMed Google Scholar * Jonathan S. Friedlaender View author publications You can
also search for this author inPubMed Google Scholar * Toomas Kivisild View author publications You can also search for this author inPubMed Google Scholar * George Koki View author
publications You can also search for this author inPubMed Google Scholar * Pradiptajati Kusuma View author publications You can also search for this author inPubMed Google Scholar * D.
Andrew Merriwether View author publications You can also search for this author inPubMed Google Scholar * Francois-X. Ricaut View author publications You can also search for this author
inPubMed Google Scholar * Joseph T. S. Wee View author publications You can also search for this author inPubMed Google Scholar * Nick Patterson View author publications You can also search
for this author inPubMed Google Scholar * Johannes Krause View author publications You can also search for this author inPubMed Google Scholar * Ron Pinhasi View author publications You can
also search for this author inPubMed Google Scholar * David Reich View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS N.P., J.K., R.P. and D.R.
supervised the study. M.S., F.V., S.B., G.R.C., and C.R. assembled archaeological material and information. P.S., C.P., Q.F., M.L., S.M., N.R. and D.R. analysed genetic data. C.P., K.Si.,
F.P., D.F., E.H., M.N., N.R, and K.St. performed laboratory work. S.A., M.P.C., F.R.F., J.S.F., T.K., G.K., P.K., D.A.M., F-X.R., and J.T.S.W. assembled the sample collection from
present-day populations. P.S. and D.R. wrote the manuscript with major input from C.P., M.S., F.V., G.R.C., M.P.C., J.S.F, J.K. and R.P. and additional input from all other co-authors.
CORRESPONDING AUTHORS Correspondence to Pontus Skoglund, Ron Pinhasi or David Reich. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ADDITIONAL
INFORMATION REVIEWER INFORMATION _Nature_ thanks P. Bellwood, C. Capelli and the other anonymous reviewer(s) for their contribution to the peer review of this work. EXTENDED DATA FIGURES AND
TABLES EXTENDED DATA FIGURE 1 ANCIENT DNA AUTHENTICITY. A, PCA performed as for Fig. 1, but with the four ancient individuals represented only by sequences that show clear evidence of
post-mortem damage (PMD score of at least 3) to remove contaminating sequences that might be present17,18. The numbers of SNPs remaining after restriction to damaged sequences is 68,450 SNPs
for I1368; 98,722 SNPs for I1369; 83,024 SNPs for I1370; and 117,023 SNPs for CP30. The lines indicate the projection of the samples when no damage-restriction is performed. The large
number of SNPs retained, and the fact that the ancient individuals cluster tightly and have the same qualitative positioning in the plot as Fig. 1, indicates that contamination did not
contribute to our findings. We also find that estimates of Papuan ancestry using PMD score restricted data are consistent with those obtained using the full data (see Methods). B,
Post-mortem damage patterns for genome-wide in-solution enrichment data from four ancient individuals. EXTENDED DATA FIGURE 2 _F_-STATISTICS DOCUMENT THE OCEANIAN ANCESTRY CLINE. A, Shared
genetic drift with the ancient Vanuatu individuals is negatively related to shared drift with Australians. Except for the ancient Tongan individual, populations from Taiwan, the Philippines
and Polynesia share the most genetic drift with the ancient Vanuatu individuals, who are not shown in the plot because they are used as reference in the computation. The trend line was
fitted without the East Asian populations in the off-cline cluster. The absence of off-cline Oceanian individuals suggests the possibility that present-day Oceanians may largely be derived
from a mixture of two source populations. B, The ancient Vanuatu individuals and the ancient Tongan individual maximize statistics of the form _f_4(Yoruba, _Test_; Australian, Oceanian),
suggesting that they are the most closely related to the East Asian ancestry in Oceanians of any sampled population. The trend line was fitted using populations >0.005 on the _x_-axis,
together with the two populations with the lowest values on the _x_-axis (Papuan and New_Guinea). C, Biplot of First Remote Oceanian ancestry proportions against conditional heterozygosity.
Populations with intermediate admixture proportion show the greatest genetic diversity. Thick and thin error bars in all panels are 1 and 1.96 standard errors of the estimate, respectively.
EXTENDED DATA FIGURE 3 ADMIXTURE DATE ESTIMATES. A, Histogram of the point estimate dates in Fig. 2d. B, Admixture date estimates for Tongans using different pairs of source populations
(‘Lapita’ in this figure refers to the pool of ancient Vanuatu individuals). Error bars show 1 (thick whiskers) and 1.96 (thin whiskers) standard errors. WGA, whole-genome amplified DNA.
EXTENDED DATA FIGURE 4 ADMIXTURE GRAPH INFERRED USING _TREEMIX_. A, A simple tree-like model without admixture fits the data poorly, as can be seen from the matrix of residuals between
empirical and modelled allele frequency covariance on the right. B, The optimal placement of a single 25% admixture event is from the lineage related to New Guinean Highlanders into the
lineage leading to Tongans. Tongans derive the other portion of their ancestry from the lineage leading to the two ancient groups of individuals. This graph has no significant deviations
between empirical and modelled allele frequency covariances. EXTENDED DATA FIGURE 5 ADMIXTURE GRAPHS MODELLING THE POPULATION HISTORY OF AUSTRALIANS. Outlier _f_4-statistics are shown (|_Z_|
> 3). A, A model with a single admixture edge positing that Australians are an outgroup to the Papuan ancestry in Tongans does not fit the data (5 outlier statistics). B, An alternative
model with 2 admixture edges in which the Papuan ancestry in Tongans also contributed to Australians fits the data (no outliers). C, A model with 2 admixture edges in which New Guinean
Highlanders are admixed from an Australian source after the divergence of the Papuan source in Tongans does not fit the data (5 outliers). D, A model with 2 admixture edges in which the
Papuan ancestry in Tongans is intermediate between the New Guinean Highlander lineage and the Australian lineage. Branch lengths are in units of _F_ST × 1,000. Lapita in this figure refers
only to Vanuatu, which is the only group for which we have multiple individuals. EXTENDED DATA FIGURE 6 FIRST REMOTE OCEANIAN ANCESTRY TODAY COMES PRIMARILY FROM FEMALES. A, Illustration of
the rationale for using the X chromosome to study asymmetrical admixture between males and females. The example on the left illustrates admixture with equal proportion of males and females
in both the red and the yellow ancestral population. The example on the right illustrates an extreme case of asymmetrical admixture where the red ancestral population only contributes
females and the yellow ancestral population only contributes males to the admixed generation, demonstrating the disproportional contribution of X chromosomes by females to the admixed
population. B, Female and male ancestral contributions based on an admixture model fitted to estimated ancestry proportions on the autosomes and X chromosome. We show the 95%, 70%, and 5%
highest posterior intervals for four selected populations from Polynesia (Samoans), the Solomon Islands (Kolombangara), Bougainville (Nasioi), and mainland New Guinea (Papuans).
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION This file contains Supplementary Notes 1-3, Supplementary Table 1 and additional references. (PDF 420 kb) POWERPOINT SLIDES POWERPOINT
SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Skoglund, P., Posth, C., Sirak,
K. _et al._ Genomic insights into the peopling of the Southwest Pacific. _Nature_ 538, 510–513 (2016). https://doi.org/10.1038/nature19844 Download citation * Received: 20 April 2016 *
Accepted: 13 September 2016 * Published: 03 October 2016 * Issue Date: 27 October 2016 * DOI: https://doi.org/10.1038/nature19844 SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative






