- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Inflammation promotes regeneration of injured tissues through poorly understood mechanisms, some of which involve interleukin (IL)-6 family members, the expression of which is
elevated in many diseases including inflammatory bowel diseases and colorectal cancer. Here we show in mice and human cells that gp130, a co-receptor for IL-6 cytokines, triggers activation
of YAP and Notch, transcriptional regulators that control tissue growth and regeneration, independently of the gp130 effector STAT3. Through YAP and Notch, intestinal gp130 signalling
stimulates epithelial cell proliferation, causes aberrant differentiation and confers resistance to mucosal erosion. gp130 associates with the related tyrosine kinases Src and Yes, which are
activated on receptor engagement to phosphorylate YAP and induce its stabilization and nuclear translocation. This signalling module is strongly activated upon mucosal injury to promote
healing and maintain barrier function. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant
access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions *
Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SPROUTY2 LIMITS INTESTINAL TUFT AND GOBLET CELL NUMBERS THROUGH GSK3Β-MEDIATED RESTRICTION OF EPITHELIAL IL-33
Article Open access 05 February 2021 LIVER X RECEPTOR UNLINKS INTESTINAL REGENERATION AND TUMORIGENESIS Article Open access 20 November 2024 EPITHELIAL GREMLIN1 DISRUPTS INTESTINAL
EPITHELIAL-MESENCHYMAL CROSSTALK TO INDUCE A WNT-DEPENDENT ECTOPIC STEM CELL NICHE THROUGH STROMAL REMODELLING Article Open access 04 June 2025 ACCESSION CODES PRIMARY ACCESSIONS
ARRAYEXPRESS * E-MTAB-2400 DATA DEPOSITS Microarray data reported here have been deposited in the ArrayExpress database under accession E-MEXP-E-MTAB-2400. REFERENCES * Ben-Neriah, Y. &
Karin, M. Inflammation meets cancer, with NF-κB as the matchmaker. _Nature Immunol._ 12, 715–723 (2011) CAS Google Scholar * Medzhitov, R. Origin and physiological roles of inflammation.
_Nature_ 454, 428–435 (2008) ADS CAS PubMed Google Scholar * Baddour, J. A., Sousounis, K. & Tsonis, P. A. Organ repair and regeneration: an overview. _Birth Defects Res. C_ 96, 1–29
(2012) CAS Google Scholar * Johnson, R. & Halder, G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. _Nature Rev. Drug Discov._ 13,
63–79 (2014) CAS Google Scholar * Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and cancer. _Cell_ 140, 883–899 (2010) CAS PubMed PubMed Central Google
Scholar * Neurath, M. F. New targets for mucosal healing and therapy in inflammatory bowel diseases. _Mucosal Immunol._ 7, 6–19 (2014) CAS PubMed Google Scholar * Garbers, C. et al.
Plasticity and cross-talk of interleukin 6-type cytokines. _Cytokine Growth Factor Rev._ 23, 85–97 (2012) CAS PubMed Google Scholar * Kishimoto, T. IL-6: from its discovery to clinical
applications. _Int. Immunol._ 22, 347–352 (2010) CAS PubMed Google Scholar * Yoshimura, A., Naka, T. & Kubo, M. SOCS proteins, cytokine signalling and immune regulation. _Nature Rev.
Immunol._ 7, 454–465 (2007) CAS Google Scholar * Putoczki, T. & Ernst, M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. _J. Leukoc. Biol._ 88,
1109–1117 (2010) CAS PubMed Google Scholar * Rose-John, S., Mitsuyama, K., Matsumoto, S., Thaiss, W. M. & Scheller, J. Interleukin-6 trans-signaling and colonic cancer associated with
inflammatory bowel disease. _Curr. Pharm. Des._ 15, 2095–2103 (2009) CAS PubMed Google Scholar * Pilati, C. et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular
adenomas. _J. Exp. Med._ 208, 1359–1366 (2011) CAS PubMed PubMed Central Google Scholar * Grivennikov, S. et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells
and development of colitis-associated cancer. _Cancer Cell_ 15, 103–113 (2009) CAS PubMed PubMed Central Google Scholar * Rebouissou, S. et al. Frequent in-frame somatic deletions
activate gp130 in inflammatory hepatocellular tumours. _Nature_ 457, 200–204 (2009) ADS CAS PubMed Google Scholar * Tanaka, T., Narazaki, M. & Kishimoto, T. Therapeutic targeting of
the interleukin-6 receptor. _Annu. Rev. Pharmacol. Toxicol._ 52, 199–219 (2012) CAS PubMed Google Scholar * Cai, J. et al. The Hippo signaling pathway restricts the oncogenic potential of
an intestinal regeneration program. _Genes Dev._ 24, 2383–2388 (2010) CAS PubMed PubMed Central Google Scholar * Okamoto, R. et al. Requirement of Notch activation during regeneration
of the intestinal epithelia. _Am. J. Physiol. Gastrointest. Liver Physiol._ 296, G23–G35 (2009) CAS PubMed Google Scholar * Yu, F. X. & Guan, K. L. The Hippo pathway: regulators and
regulations. _Genes Dev._ 27, 355–371 (2013) CAS PubMed PubMed Central Google Scholar * Rosenbluh, J. et al. β-Catenin-driven cancers require a YAP1 transcriptional complex for survival
and tumorigenesis. _Cell_ 151, 1457–1473 (2012) CAS PubMed PubMed Central Google Scholar * Bray, S. J. Notch signalling: a simple pathway becomes complex. _Nature Rev. Mol. Cell Biol._
7, 678–689 (2006) CAS Google Scholar * Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum,
cecum) axes of the intestine. _J. Biol. Chem._ 277, 33275–33283 (2002) CAS PubMed Google Scholar * Camargo, F. D. et al. YAP1 increases organ size and expands undifferentiated progenitor
cells. _Curr. Biol._ 17, 2054–2060 (2007) CAS PubMed Google Scholar * Zhou, D. et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis
by inhibition of Yes-associated protein (Yap) overabundance. _Proc. Natl Acad. Sci. USA_ 108, E1312–E1320 (2011) CAS PubMed Google Scholar * Fre, S. et al. Notch signals control the fate
of immature progenitor cells in the intestine. _Nature_ 435, 964–968 (2005) ADS CAS PubMed Google Scholar * van Es, J. H. et al. Notch/γ-secretase inhibition turns proliferative cells in
intestinal crypts and adenomas into goblet cells. _Nature_ 435, 959–963 (2005) ADS CAS PubMed Google Scholar * Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single
intestinal stem cell: mechanism and applications. _Science_ 340, 1190–1194 (2013) ADS CAS PubMed Google Scholar * Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells
in intestinal crypts. _Nature_ 469, 415–418 (2011) ADS CAS PubMed Google Scholar * Murakami, D. et al. Presenilin-dependent gamma-secretase activity mediates the intramembranous cleavage
of CD44. _Oncogene_ 22, 1511–1516 (2003) CAS PubMed Google Scholar * Zhang, J. et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo
pathway. _Nature Cell Biol._ 11 1444–1450 10.1038/ncb1993 (2009) CAS PubMed Google Scholar * Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity
to distinguish mouse trophectoderm from inner cell mass. _Dev. Cell_ 16, 398–410 (2009) CAS Google Scholar * Chen, J., Elfiky, A., Han, M., Chen, C. & Saif, M. W. The role of Src in
colon cancer and its therapeutic implications. _Clin. Colorectal Cancer_ 13, 5–13 (2014) PubMed Google Scholar * Levy, D., Adamovich, Y., Reuven, N. & Shaul, Y. Yap1 phosphorylation by
c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. _Mol. Cell_ 29, 350–361 (2008) CAS PubMed Google Scholar * Azzolin, L. et al. YAP/TAZ
incorporation in the β-catenin destruction complex orchestrates the Wnt response. _Cell_ 158, 157–170 (2014) CAS PubMed Google Scholar * Sudol, M. Yes-associated protein (YAP65) is a
proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. _Oncogene_ 9, 2145–2152 (1994) CAS PubMed Google Scholar * Tsutsumi, R. et al. YAP and TAZ,
Hippo signaling targets, act as a rheostat for nuclear SHP2 function. _Dev. Cell_ 26, 658–665 (2013) CAS PubMed Google Scholar * Tschaharganeh, D. F. et al. Yes-associated protein
up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. _Gastroenterology_ 144, 1530–1542 (2013) CAS PubMed PubMed Central Google Scholar * Li, Y.,
Hibbs, M. A., Gard, A. L., Shylo, N. A. & Yun, K. Genome-wide analysis of N1ICD/RBPJ targets _in vivo_ reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors
by Notch1. _Stem Cells_ 30, 741–752 (2012) PubMed PubMed Central Google Scholar * Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. _Cell_ 150,
780–791 (2012) CAS PubMed PubMed Central Google Scholar * Gordon, M. et al. The tumor suppressor gene, RASSF1A, is essential for protection against inflammation-induced injury. _PLoS
ONE_ 8, e75483 (2013) ADS CAS PubMed PubMed Central Google Scholar * Tamm, C., Bower, N. & Anneren, C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2
signaling pathway downstream of LIF. _J. Cell Sci._ 124, 1136–1144 (2011) PubMed Google Scholar * Raz, R., Lee, C. K., Cannizzaro, L. A., d’Eustachio, P. & Levy, D. E. Essential role
of STAT3 for embryonic stem cell pluripotency. _Proc. Natl Acad. Sci. USA_ 96, 2846–2851 (1999) ADS CAS PubMed Google Scholar * Taniguchi, K. & Karin, M. IL-6 and related cytokines
as the critical lynchpins between inflammation and cancer. _Semin. Immunol._ 26, 54–74 (2014) CAS PubMed Google Scholar * Hedvat, M. et al. The JAK2 inhibitor AZD1480 potently blocks
Stat3 signaling and oncogenesis in solid tumors. _Cancer Cell_ 16, 487–497 (2009) CAS PubMed PubMed Central Google Scholar * Cordero, J. B. et al. c-Src drives intestinal regeneration
and transformation. _EMBO J._ 33, 1474–1491 (2014) CAS PubMed PubMed Central Google Scholar * Takeda, K. et al. Enhanced Th1 activity and development of chronic enterocolitis in mice
devoid of Stat3 in macrophages and neutrophils. _Immunity_ 10, 39–49 (1999) CAS PubMed Google Scholar * Zhang, N. et al. The Merlin/NF2 tumor suppressor functions through the YAP
oncoprotein to regulate tissue homeostasis in mammals. _Dev. Cell_ 19, 27–38 (2010) CAS PubMed PubMed Central Google Scholar * Ootani, A. et al. Sustained _in vitro_ intestinal
epithelial culture within a Wnt-dependent stem cell niche. _Nature Med._ 15, 701–706 (2009) CAS PubMed Google Scholar * Holzer, R. G. et al. Saturated fatty acids induce c-Src clustering
within membrane subdomains, leading to JNK activation. _Cell_ 147, 173–184 (2011) CAS PubMed PubMed Central Google Scholar * Hu, H. T. et al. Tie2-R849W mutant in venous malformations
chronically activates a functional STAT1 to modulate gene expression. _J. Invest. Dermatol._ 128 2325–2333 10.1038/jid.2008.89 (2008) CAS PubMed Google Scholar * Nam, J. S., Ino, Y.,
Sakamoto, M. & Hirohashi, S. Src family kinase inhibitor PP2 restores the E-cadherin/catenin cell adhesion system in human cancer cells and reduces cancer metastasis. _Clin. Cancer Res._
8, 2430–2436 (2002) CAS PubMed Google Scholar * Lee, S. H. et al. ERK activation drives intestinal tumorigenesis in _Apc__min/+_ mice. _Nature Med._ 16, 665–670 (2010) CAS PubMed
Google Scholar * Hu, L., Zaloudek, C., Mills, G. B., Gray, J. & Jaffe, R. B. _In vivo_ and _in vitro_ ovarian carcinoma growth inhibition by a phosphatidylinositol 3-kinase inhibitor
(LY294002). _Clin. Cancer Res._ 6, 880–886 (2000) CAS PubMed Google Scholar * Katakura, K. et al. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. _J.
Clin. Invest._ 115, 695–702 (2005) CAS PubMed PubMed Central Google Scholar * Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche.
_Nature_ 459, 262–265 (2009) ADS CAS PubMed Google Scholar * Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's
epithelium. _Gastroenterology_ 141, 1762–1772 (2011) CAS PubMed Google Scholar * Guma, M. et al. Constitutive intestinal NF-κB does not trigger destructive inflammation unless accompanied
by MAPK activation. _J. Exp. Med._ 208, 1889–1900 (2011) CAS PubMed PubMed Central Google Scholar * Barker, N. et al. Identification of stem cells in small intestine and colon by marker
gene Lgr5. _Nature_ 449, 1003–1007 (2007) ADS CAS PubMed Google Scholar * Umemura, A. et al. Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition.
_Cell Metab._ 20, 133–144 (2014) CAS PubMed PubMed Central Google Scholar * Mitchell, C. & Willenbring, H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in
mice. _Nature Protocols_ 3, 1167–1170 (2008) CAS PubMed Google Scholar * Kozak, I. et al. A degenerative retinal process in HIV-associated non-infectious retinopathy. _PLoS ONE_ 8, e74712
(2013) ADS CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank D. Pan and S. Akira for _Yap__fl/fl_ and _Stat3__fl/fl_ mice, respectively. We also
thank D. L. Gumucio for a plasmid containing the 12.4-kb villin promoter, T. Sato, H. Clevers and Y. Hippo for protocols describing intestinal organoid culture, C. Kuo for
R-spondin1-producing cells, D. Huszar for AZD1480, F. Schaper for plasmids, L. Eckmann for advice, A. Umemura, H. Nakagawa, H. Ogata, E. J. Park, G. Y. Yu, J. Font-Burgada, D. Dhar, J. Kim
and E. Seki for providing liver samples, J. Zhao, T. Meerloo, Y. Jones, L. Gapuz, R. Ly, N. Varki, D. Aki, N. Hiramatsu, T. Moroishi, Y. Endo, H. Nishinakamura, A. Chang and T. Lee for
technical advice and assistance, and Cell Signaling, Santa Cruz Biotechnology and GeneTex for antibodies. This work was supported by Postdoctoral Fellowship for Research Abroad and Research
Fellowship for Young Scientists from the Japan Society for the Promotion of Science, a Uehara Memorial Foundation Fellowship, the Mochida Memorial Foundation for Medical and Pharmaceutical
Research, and the Kanae Foundation for the Promotion of Medical Science to K.T.; a traveling grant NSC-101-2918-I-006-005 and a research grant NSC-103-2320-B-006-032 by National Science
Council of Taiwan to L.-W.W.; NIH R00DK088589, FCCC-Temple University Nodal grant, AACR-Landon Innovator Award in Tumor Microenvironment, and the Pew Scholar in Biomedical Sciences Program
for S.I.G.; a CCFA fellowship (RFA2927) to P.R.d.J.; Croucher Foundation and China Postdoctoral Science Foundation to K.W.; by the Research Service of the Department of Veterans Affairs to
S.B.H.; by the NIH and the UCSD Digestive Disease Research Center Grant to J.T.C. and W.J.S.; by the NIH EY022611 and CA132809 to K.-L.G.; and by the NIH CA118165-09 and AACR to M.K., who is
an American Cancer Society Research Professor and holds the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory
of Gene Regulation and Signal Transduction, University of California, San Diego, La Jolla, California 92093, USA, Koji Taniguchi, Li-Wha Wu, Sergei I. Grivennikov, Kepeng Wang & Michael
Karin * Departments of Pharmacology and Pathology, University of California, San Diego, La Jolla, California 92093, USA, Koji Taniguchi, Ian Lian, Fa-Xing Yu, Kun-Liang Guan & Michael
Karin * Department of Surgery and Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan, Koji Taniguchi & Yoshihiko Maehara * Department of
Microbiology and Immunology, Keio University School of Medicine, Tokyo 160-8582, Japan, Koji Taniguchi & Akihiko Yoshimura * Institute of Molecular Medicine, College of Medicine,
National Cheng Kung University, Tainan 70101, Taiwan, Li-Wha Wu * Fox Chase Cancer Center, Cancer Prevention and Control Program, Philadelphia, 19111, Pennsylvania, USA Sergei I. Grivennikov
* Department of Medicine, University of California, San Diego, La Jolla, California 92093, USA, Petrus R. de Jong & Eyal Raz * Moores Cancer Center, University of California, San Diego,
La Jolla, California 92093, USA, Ian Lian, Fa-Xing Yu, Kun-Liang Guan & Michael Karin * Department of Biology, Lamar University, PO Box 10037, Beaumont, Texas 77710, USA, Ian Lian *
Children’s Hospital and Institutes of Biomedical Sciences, Fudan University, Shanghai, 200032, China Fa-Xing Yu * Department of Medicine, VA San Diego Healthcare System, San Diego, 92161,
California, USA Samuel B. Ho * Division of Gastroenterology, Department of Medicine, Inflammatory Bowel Disease Center, School of Medicine, University of California, San Diego, La Jolla,
California 92093, USA, Brigid S. Boland, John T. Chang & William J. Sandborn * Department of Medicine, Medical University of South Carolina, Charleston, 29425, South Carolina, USA Gary
Hardiman * CSRC and BIMRC, San Diego State University, San Diego, 92182, California, USA Gary Hardiman * Japan Science and Technology Agency, CREST, Tokyo 102-0076, Japan, Akihiko Yoshimura
* Inserm, UMR 1162, Génomique fonctionnelle des tumeurs solides, IUH, Paris 75010, France, Jessica Zucman-Rossi * Université Paris Descartes, Labex Immuno-oncology, Sorbonne Paris Cité,
Faculté de Medicine, Paris 75006, France, Jessica Zucman-Rossi Authors * Koji Taniguchi View author publications You can also search for this author inPubMed Google Scholar * Li-Wha Wu View
author publications You can also search for this author inPubMed Google Scholar * Sergei I. Grivennikov View author publications You can also search for this author inPubMed Google Scholar *
Petrus R. de Jong View author publications You can also search for this author inPubMed Google Scholar * Ian Lian View author publications You can also search for this author inPubMed
Google Scholar * Fa-Xing Yu View author publications You can also search for this author inPubMed Google Scholar * Kepeng Wang View author publications You can also search for this author
inPubMed Google Scholar * Samuel B. Ho View author publications You can also search for this author inPubMed Google Scholar * Brigid S. Boland View author publications You can also search
for this author inPubMed Google Scholar * John T. Chang View author publications You can also search for this author inPubMed Google Scholar * William J. Sandborn View author publications
You can also search for this author inPubMed Google Scholar * Gary Hardiman View author publications You can also search for this author inPubMed Google Scholar * Eyal Raz View author
publications You can also search for this author inPubMed Google Scholar * Yoshihiko Maehara View author publications You can also search for this author inPubMed Google Scholar * Akihiko
Yoshimura View author publications You can also search for this author inPubMed Google Scholar * Jessica Zucman-Rossi View author publications You can also search for this author inPubMed
Google Scholar * Kun-Liang Guan View author publications You can also search for this author inPubMed Google Scholar * Michael Karin View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS K.T. and M.K. conceived the project. K.T., L.-W.W., S.I.G., P.R.d.J., I.L., F.-X.Y., K.W., G.H. performed the experiments. K.T., L.-W.W.,
P.R.d.J, G.H. and M.K. analysed data. J.Z.-R. provided gp130 mutants, S.B.H., J.T.C., B.S.B. and W.J.S. provided human specimens, S.I.G., E.R., Y.M., A.Y., J.Z.-R. and K.-L.G. provided
conceptual advice. K.T., L.-W.W. and M.K. wrote the manuscript, with all authors contributing to the writing and providing advice. CORRESPONDING AUTHOR Correspondence to Michael Karin.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIGURE 1 GP130ACT EXPRESSION AND INTESTINAL
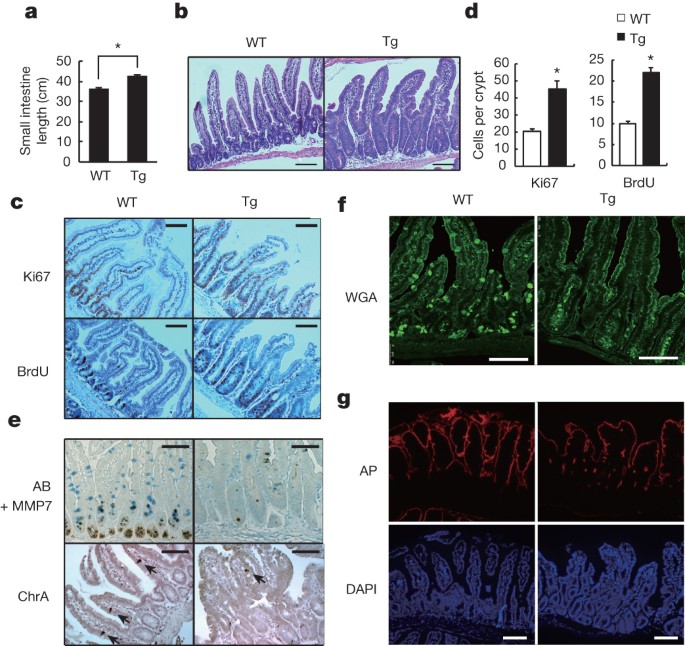
PHENOTYPE. A, Schematic diagram of the _villin-gp130__Act_ transgenic (Tg) construct and the gp130Act and gp130SA variants. B, Expression of _gp130__Act_ in the _villin-gp130__Act_ jejunum
was confirmed by RT–PCR with specific primers for human _gp130_. Cyclophilin (_CPH_) was used as an internal control. C, Representative images of wild-type (WT) and _villin-gp130__Act_
intestines. c-Myc and cyclin D1 (D) and TUNEL (red, TUNEL; blue, DAPI) (E) staining of paraffin-embedded sections of wild-type and _villin-gp130__Act_ small intestines from 3-month-old mice.
Positive cells were enumerated in each villus or crypt (_n_ = 6). Data represent averages ± s.d.; _P_ < 0.05. F, MMP7-, AB- and ChrA-positive cells in wild-type and _villin-gp130__Act_
small intestines were enumerated in each villus or crypt (_n_ = 6). Data represent averages ± s.d.; _P_ < 0.05. G, Paraffin-embedded sections of wild-type and _villin-gp130__Act_ small
intestines were analysed by PAS and lysozyme staining. Positive cells were enumerated in each villus or crypt (_n_ = 6). Data represent averages ± s.d.; _P_ < 0.05. H, Cryptdin mRNA in
wild-type and _villin-gp130__Act_ jejunum was detected by _in situ_ hybridization. I, Transmission electron microscopy (TEM) of the apical surface of wild-type and transgenic small
intestines. Scale bars represent 100 μm (D, E, G, H) and 1 μm (I) and all data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 2 ABERRANT INTESTINAL
DIFFERENTIATION AND ACTIVATION OF GP130 EFFECTORS IN _GP130__ACT_ MICE. A, Paraffin-embedded sections of wild-type and _villin-gp130__Act_ (Tg) small intestines were analysed by CD45
staining. B, Lysates of wild-type and _villin-gp130__Act_ jejuna were prepared, and expression of _Il6_ and _Tnf_ mRNAs was analysed by qRT–PCR (_n_ = 3). Results are averages ± s.e.m.; _P_
< 0.05. C, H&E and PAS staining of paraffin-embedded sections of wild-type and _villin-gp130__Act_ large intestines. D, pSTAT1, pERK1/2, pS6 and CD44 C-terminal stainings of
paraffin-embedded sections of wild-type and _villin-gp130__Act_ small intestines. Positive cells were enumerated in each villus or crypt (_n_ = 4). Data represent averages ± s.d.; _P_ <
0.05. E, YAP, Ki67 and pSTAT3 stainings of paraffin-embedded sections of wild-type and _villin-gp130__Act_ large intestines. Positive cells were enumerated in each crypt (_n_ = 4). Data
represent averages ± s.d.; _P_ < 0.05. Scale bars represent 100 μm (A, C-E) and all data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 3 IL-6 AND GP130
INDUCE NOTCH AND YAP ACTIVATION IN INTESTINAL ORGANOIDS AND CANCER CELLS, AND GENE EXPRESSION ANALYSIS OF INTESTINAL CRYPTS. A, B, Wild-type and _villin-gp130__Act_ organoids were cultured,
their RNA extracted, and expression of the indicated mRNA species was measured by qRT–PCR (_n_ = 3). Results are averages ± s.e.m.; _P_ < 0.05. C, Appearance of wild-type and
_villin-gp130__Act_ small intestinal organoids cultured in standard EGF/Noggin/R-spondin 1 medium. D, Nuclei of T84 colon cancer cells transfected with either empty vector (EV) or a vector
encoding superactive gp130 (gp130SA) were lysed and subjected to immunoblot analysis with the indicated antibodies. Lamin A, a nuclear marker. Actin, a loading control. E, Lysates of
serum-starved SW480 (upper) or DLD1 (lower) colon cancer cells treated for 0–480 min with IL-6 at 50 ng ml−1 were subjected to immunoblot analysis using the indicated antibodies. F, Nuclei
of serum-starved HT29 colon cancer cells treated for 24 h with IL-6 at 0–50 ng ml−1 were lysed and subjected to immunoblot analysis with the indicated antibodies. HDAC, a nuclear marker and
loading control. G, Lysates of primary mouse hepatocytes treated for 0–120 min with IL-6 at 50 ng ml−1 were subjected to immunoblot analysis using the indicated antibodies. H, Microarray
analysis was performed using the Illumina MouseWG-6 v2 Expression BeadChip on RNA extracted from wild-type and _villin-gp130__Act_ small intestinal crypts (_n_ = 3 per group). Data were
normalized and analysed as described and expression of the indicated genes is shown as fold-induction compared to wild-type crypts. I, RNA was extracted from wild-type and
_villin-gp130__Act_ small intestinal organoids, and _Areg_ mRNA expression was measured by qRT–PCR (_n_ = 3). Results are averages ± s.e.m.; _P_ < 0.05. Scale bars represent 100 μm (C)
and all data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 4 ABERRANT INTESTINAL DIFFERENTIATION IN _GP130__ACT_ MICE DEPENDS ON NOTCH AND YAP BUT NOT ON
STAT3. A, MMP7 staining of paraffin-embedded sections of control and DBZ-treated (10 μmol kg−1) _villin-gp130__Act_ small intestines. Positive cells were enumerated in each crypt (_n_ = 3).
Data represent averages ± s.d.; _P_ < 0.05. B, MMP7 and lysozyme staining of paraffin-embedded sections of _villin-gp130__Act_ and _villin-gp130__Act__/Yap__ΔIEC_ small intestines.
Positive cells were enumerated in each crypt (_n_ = 4). Data represent averages ± s.d.; _P_ < 0.05. C, PAS, Ki67, YAP, pSTAT3, HES1 and MMP7 staining of paraffin-embedded sections of
_villin-gp130__Act_ and _villin-gp130__Act__/Stat3__ΔIEC_ small intestines. Positive cells were enumerated in each villus or crypt (_n_ = 4). Data represent averages ± s.d.; _P_ < 0.05.
Scale bars represent 100 μm (A-C) and all data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 5 MEK AND PI(3)K INHIBITORS HAVE NO EFFECT ON ABERRANT
INTESTINAL HOMEOSTASIS IN _GP130__ACT_ MICE. A, PAS, Ki67, YAP and pERK1/2 staining of paraffin-embedded sections of control and PD0325901-treated (25 mg kg−1) _villin-gp130__Act_ small
intestines. Positive cells were enumerated in each villus or crypt (_n_ = 3). Data represent averages ± s.d.; _P_ < 0.05. B, PAS, Ki67, YAP and pS6 staining of paraffin-embedded sections
of control and LY294002-treated (100 mg kg−1) _villin-gp130__Act_ small intestines. Positive cells were enumerated in each villus or crypt (_n_ = 3). Data represent averages ± s.d.; _P_ <
0.05. Scale bars represent 100 μm (A, B). EXTENDED DATA FIGURE 6 GP130 ACTIVATES YAP VIA A HIPPO-INDEPENDENT BUT TYROSINE PHOSPHORYLATION-DEPENDENT MECHANISM, AND GP130 INTERACTS WITH SRC
AND YES. A, Lysates of wild-type and _villin-gp130__Act_ jejuna, which are the same as the ones in Fig. 2a, were analysed for expression and phosphorylation of the indicated proteins. B,
Lysates of HT29 colon cancer cells transfected with either empty vector (EV), active gp130 (gp130Act), or superactive gp130 (gp130SA) were subjected to immunoblot analysis with pSrc (Y419),
total Src and GAPDH antibodies. GAPDH, a loading control. C, Lysates of HCA7 colon cancer cells infected with EV, wild-type gp130, gp130Act, or gp130SA lentiviruses were immunoprecipitated
with anti-YAP antibody and blotted with the indicated antibodies. D, Lysates of HT29 colon cancer cells infected with EV, wild-type gp130, gp130Act, or gp130SA lentiviruses were immunoblot
analysed for expression and phosphorylation of the indicated proteins. E, Serum-starved HCT116 cells were stimulated with 10% FBS, IL-6 (100 ng ml−1), or IL-11 (100 ng ml−1) for 30 min.
Total cell lysates were analysed for expression and phosphorylation of the indicated proteins. F, Left: pSrc and YAP staining of livers from untreated wild-type mice (control) and wild-type
mice 48 h after partial hepatectomy (PH). Scale bars represent 100 µm. Middle: lysates of livers from control mice and mice 48 h after partial hepatectomy were subjected to immunoblot
analysis with the indicated antibodies. Right: lysates of livers from vehicle (DMSO)-treated and PP2-treated mice 48 h after partial hepatectomy were subjected to immunoblot analysis with
the indicated antibodies. G, Top: HEK293T cells were transfected with plasmids expressing Flag–YAP. Twenty-four hours after transfection, the cells were pre-treated for 1 h with 0.1% DMSO
(vehicle control), PP2 (10 μM) or AZD1480 (1 μM) and then were treated with 50 μg ml−1 cycloheximide for different time points. Total cell lysates were subjected to immunoblot analysis with
the indicated antibodies. Bottom: HEK293T cells were transfected with Flag–YAP as above. Twenty-four hours after transfection, the cells were pre-treated for 1 h with 0.1% DMSO (vehicle
control), AZD0530 (10 μM) or SU6656 (10 μM) and then were treated with 50 μg ml−1 cycloheximide for different time points. Total cell lysates were analysed as above. H, HEK293T cells were
transfected with either empty or constitutively active (CA) Src expression vectors. After 48 h, the cells were lysed and expression of the indicated proteins determined by immunoblot
analysis. I, HT29 cells were collected with or without 10 ng ml−1 IL-6 stimulation for 2 h. Lysates were analysed by immunoblot with the indicated antibodies. These are the loading controls
for the data shown in Fig. 4e. J, HEK293T cells were transfected with expression vectors encoding Src and Flag-tagged gp130Act, Flag-tagged gp130Act (Δ771–811), Flag-tagged gp130Act
(Δ812–827) or empty vector. Cells were collected 48 h later. Cell lysates were immunoprecipitated with Flag antibody and analysed by immunoblot with the indicated antibodies. K, Nuclei of
T84 colon cancer cells infected with EV, gp130Act, gp130Act (Δ771–811) or gp130Act (Δ812–827) lentiviruses were prepared and subjected to immunoblot analysis with the indicated antibodies.
HDAC1, a nuclear marker and loading control. All data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 7 SFK ACTIVITY IS REQUIRED FOR YAP ACTIVATION. A,
Transgenic (Tg) mice (_n_ = 4 per group) were treated with PP2 (5 mg kg−1) or vehicle once a day for 5 days. Small intestines were isolated, sectioned and stained as indicated. Positive
cells were enumerated in each villus or crypt. Data represent averages ± s.d.; _P_ < 0.05. B, C, Wild-type and _villin-gp130__Act_ small intestinal organoids were treated with DMSO,
AZD0530 (10 μM) (B), AZD1480 (1 μM) or DBZ (10 μM) (C) for 24 h, stained with YAP antibody and counter stained with DAPI and photographed under a fluorescent microscope. D, Wild-type and
_villin-gp130__Act_ small intestinal organoids were treated with DMSO, PP2 (10 μM) and AZD1480 (1 μM) for 24 h. Total cell lysates were subjected to immunoblot analysis with the indicated
antibodies. E, Serum-starved HT29 cells were pre-treated for 1 h with 0.1% DMSO (vehicle control), AZD1480 (10 μM) or PP2 (20 μM) before IL-6 (10 ng ml−1) stimulation for 24 h. Nuclear
extracts of HT29 cells treated without or with IL-6 in the absence or presence of AZD1480 or PP2 were subjected to immunoblot analysis with the indicated antibodies. Lamin A, a nuclear
marker; Actin, a loading control. F, Wild-type and _villin-gp130__Act_ small intestinal organoids were treated with DMSO and AZD0530 (10 μM) for 24 h. Total cell lysates were subjected to
immunoblot analysis with the indicated antibodies. Scale bars represent 100 μm (A–C). All data are representative of at least 2–3 independent experiments. EXTENDED DATA FIGURE 8 GP130ACT
CONFERS DSS RESISTANCE, INDUCES NOTCH RECEPTORS AND LIGANDS AND IMPROVES BARRIER FUNCTION. A, Left: representative images of wild-type and _villin-gp130__Act_ large intestines taken 10 days
after 3.0% DSS treatment. Right: colon length of wild-type and _villin-gp130__Act_ mice before and after DSS treatment (before: _n_ = 5 per group, after: _n_ = 4 per group). Results are
averages ± s.e.m.; _P_ < 0.05. B, Representative images of H&E stained paraffin-embedded colon sections prepared 10 days after DSS challenge of wild-type and transgenic mice as
described in Fig. 5a. Scale bars: 100 µm. C, Ki67 (left) and cleaved-caspase 3 (right) stainings were performed on paraffin-embedded colon sections from wild-type and transgenic mice at day
0 and 3 (Ki67) or 5 (cleaved-caspase 3) after 3.0% DSS treatment. D, Lysates of wild-type, _villin-gp130__Act_, _villin-gp130__Act__/Yap__ΔIEC_ and _villin-gp130__Act__/Stat3__ΔIEC_ colons
were prepared, RNA was extracted and expression of the indicated mRNA species was analysed by qRT–PCR (_n_ = 3 per group). Results are averages ± s.e.m.; _P_ < 0.05. E, F, Gut barrier
function in wild-type and _villin-gp130__Act_ mice was examined by measurements of fecal albumin (WT, _n_ = 6; Tg, _n_ = 7) (E) and FITC-Dextran translocation to blood 4 h after oral gavage
(WT, _n_ = 5; Tg, _n_ = 4) (F). Results are averages ± s.e.m.; _P_ < 0.05. G, TEM images of intestinal mucosa epithelial cell–cell junctions in wild-type and _villin-gp130__Act_ small
intestines. H, C57BL/6 mice were given regular water or 2.5% DSS for 7 days. Colonic RNA was extracted on day 10, and expression of the indicated genes was analysed by qRT–PCR (_n_ = 4).
Results are averages ± s.e.m.; _P_ < 0.05. I, Wild-type mice were given 2.5% DSS. Colonic lysates were prepared when indicated and immunoblot analysed for protein expression and
phosphorylation. J, Colon length of control and PP2-injected C57BL/6 mice after DSS treatment (_n_ = 6 per group). Results are averages ± s.e.m.; _P_ < 0.05. Scale bars represent 100 μm
(B, C) and 500 nm (G). EXTENDED DATA FIGURE 9 ENHANCED MUCOSAL REGENERATION IN _GP130__ACT_ MICE DEPENDS ON YAP AND STAT3 BUT THE TWO EFFECTORS CONTROL DIFFERENT GENES, AND YAP IS REQUIRED
FOR _IN VITRO_ SCRATCH CLOSURE. A, Left: body weight curves of DSS-treated _Yap__ΔIEC_ (squares, _n_ = 6) and _villin-gp130__Act__/Yap__ΔIEC_ (circles, _n_ = 4) mice. Results are averages ±
s.d.; _P_ < 0.05. Colon mucosal histology of _Yap__ΔIEC_ (squares, _n_ = 6) and _villin-gp130__Act__/Yap__ΔIEC_ (circles, _n_ = 4) mice was examined by H&E staining and scored 9 days
after 2.0% DSS challenge by an observer blinded to the mouse genotype. Results are averages ± s.e.m.; _P_ < 0.05. Right: body weight curves of DSS-treated _Stat3__ΔIEC_ (squares) and
_villin-gp130__Act__/Stat3__ΔIEC_ (circles) mice (_n_ = 4 per group). Results are averages ± s.d.; _P_ < 0.05. Mucosal histology of _Stat3__ΔIEC_ and _villin-gp130__Act__/Stat3__ΔIEC_
mice (_n_ = 4 per group) was examined and scored 8 days after 2.0% DSS challenge as above. Results are averages ± s.e.m.; _P_ < 0.05. B, RNA was extracted from _Yap__ΔIEC_ and
_villin-gp130__Act__/Yap__ΔIEC_ (_n_ = 3 per group) or _Stat3__ΔIEC_ and _villin-gp130__Act__/Stat3__ΔIEC_ (_n_ = 4 per group) colons, and expression of the indicated mRNA species was
measured by qRT–PCR. Results are averages ± s.e.m.; _P_ < 0.05. C, IEC6 rat intestinal epithelial cells infected with EV or gp130Act lentiviruses were grown to confluence and starved
overnight, and the monolayers were wounded by scratching and treated with DMSO, PP2 (10 μM), AZD1480 (1 μM), verteporfin (1 μg ml−1) or DBZ (10 μM). The per cent wounded area was calculated
by measuring wound closure over time (0 and 24 h). Results are averages ± s.e.m.; _P_ < 0.05 (_n_ = 5). D, Total cell lysates of IEC6 cells infected with EV + shluc (control), gp130Act +
shluc or gp130Act + shYAP lentiviruses were prepared and subjected to immunoblot analysis with the indicated antibodies. E, IEC6 cells infected with EV + shluc (control), gp130Act + shluc or
gp130Act + shYAP lentiviruses were grown to confluence and starved overnight, and the monolayers were wounded by scratching. The per cent wounded area was calculated by measuring wound
closure over time (0 and 24 h) (_n_ = 5). Results are averages ± s.e.m.; _P_ < 0.05. F, Schematic representation of the gp130–SFK–YAP–Notch pathway and its function in the injured
intestinal epithelium. Scale bars represent 100 μm (A). POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4
POWERPOINT SLIDE FOR FIG. 5 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Taniguchi, K., Wu, LW., Grivennikov, S. _et al._ A gp130–Src–YAP module links
inflammation to epithelial regeneration. _Nature_ 519, 57–62 (2015). https://doi.org/10.1038/nature14228 Download citation * Received: 07 April 2014 * Accepted: 09 January 2015 * Published:
25 February 2015 * Issue Date: 05 March 2015 * DOI: https://doi.org/10.1038/nature14228 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
:max_bytes(150000):strip_icc():focal(319x0:321x2)/people_social_image-60e0c8af9eb14624a5b55f2c29dbe25b.png)