- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
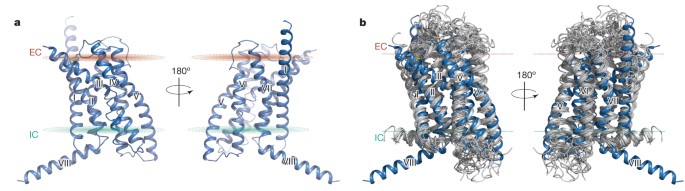
ABSTRACT Binding of the glucagon peptide to the glucagon receptor (GCGR) triggers the release of glucose from the liver during fasting; thus GCGR plays an important role in glucose
homeostasis. Here we report the crystal structure of the seven transmembrane helical domain of human GCGR at 3.4 Å resolution, complemented by extensive site-specific mutagenesis, and a
hybrid model of glucagon bound to GCGR to understand the molecular recognition of the receptor for its native ligand. Beyond the shared seven transmembrane fold, the GCGR transmembrane
domain deviates from class A G-protein-coupled receptors with a large ligand-binding pocket and the first transmembrane helix having a ‘stalk’ region that extends three alpha-helical turns
above the plane of the membrane. The stalk positions the extracellular domain (∼12 kilodaltons) relative to the membrane to form the glucagon-binding site that captures the peptide and
facilitates the insertion of glucagon’s amino terminus into the seven transmembrane domain. Access through your institution Buy or subscribe This is a preview of subscription content, access
via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy
this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: *
Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MOLECULAR FEATURES OF THE LIGAND-FREE GLP-1R, GCGR AND GIPR
IN COMPLEX WITH GS PROTEINS Article Open access 13 February 2024 A UNIQUE HORMONAL RECOGNITION FEATURE OF THE HUMAN GLUCAGON-LIKE PEPTIDE-2 RECEPTOR Article Open access 25 November 2020
DYNAMICS OF GLP-1R PEPTIDE AGONIST ENGAGEMENT ARE CORRELATED WITH KINETICS OF G PROTEIN ACTIVATION Article Open access 10 January 2022 ACCESSION CODES ACCESSIONS PROTEIN DATA BANK * 4L6R
DATA DEPOSITS The coordinates and the structure factors have been deposited in the Protein Data Bank under the accession code 4L6R. REFERENCES * Lagerström, M. C. & Schioth, H. B.
Structural diversity of G protein-coupled receptors and significance for drug discovery. _Nature Rev. Drug Discov._ 7, 339–357 (2008) Article Google Scholar * Cho, Y. M., Merchant, C. E.
& Kieffer, T. J. Targeting the glucagon receptor family for diabetes and obesity therapy. _Pharmacol. Ther._ 135, 247–278 (2012) Article CAS PubMed Google Scholar * Katritch, V.,
Cherezov, V. & Stevens, R. C. Structure-function of the G protein-coupled receptor superfamily. _Annu. Rev. Pharmacol. Toxicol._ 53, 531–556 (2013) Article CAS PubMed Google Scholar
* Hoare, S. R. Mechanisms of peptide and nonpeptide ligand binding to class B G-protein-coupled receptors. _Drug Discov. Today_ 10, 417–427 (2005) Article CAS PubMed Google Scholar *
Pal, K., Melcher, K. & Xu, H. E. Structure and mechanism for recognition of peptide hormones by Class B G-protein-coupled receptors. _Acta Pharmacol. Sin._ 33, 300–311 (2012) Article
CAS PubMed PubMed Central Google Scholar * Koth, C. M. et al. Molecular basis for negative regulation of the glucagon receptor. _Proc. Natl Acad. Sci. USA_ 109, 14393–14398 (2012)
Article CAS ADS PubMed PubMed Central Google Scholar * Parthier, C., Reedtz-Runge, S., Rudolph, R. & Stubbs, M. T. Passing the baton in class B GPCRs: peptide hormone activation
via helix induction? _Trends Biochem. Sci._ 34, 303–310 (2009) Article CAS PubMed Google Scholar * Underwood, C. R. et al. Crystal structure of glucagon-like peptide-1 in complex with
the extracellular domain of the glucagon-like peptide-1 receptor. _J. Biol. Chem._ 285, 723–730 (2010) Article CAS PubMed Google Scholar * Yaqub, T. et al. Identification of determinants
of glucose-dependent insulinotropic polypeptide receptor that interact with N-terminal biologically active region of the natural ligand. _Mol. Pharmacol._ 77, 547–558 (2010) Article CAS
PubMed Google Scholar * Miller, L. J., Dong, M., Harikumar, K. G. & Gao, F. Structural basis of natural ligand binding and activation of the Class II G-protein-coupled secretin
receptor. _Biochem. Soc. Trans._ 35, 709–712 (2007) Article CAS PubMed Google Scholar * Dong, M. et al. Mapping spatial approximations between the amino terminus of secretin and each of
the extracellular loops of its receptor using cysteine trapping. _FASEB J._ 26, 5092–5105 (2012) Article CAS PubMed PubMed Central Google Scholar * Miller, L. J. et al. Refinement of
glucagon-like peptide 1 docking to its intact receptor using mid-region photolabile probes and molecular modeling. _J. Biol. Chem._ 286, 15895–15907 (2011) Article CAS PubMed PubMed
Central Google Scholar * Dong, M. et al. Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore. _J. Biol.
Chem._ 286, 23888–23899 (2011) Article CAS PubMed PubMed Central Google Scholar * Gensure, R. C., Shimizu, N., Tsang, J. & Gardella, T. J. Identification of a contact site for
residue 19 of parathyroid hormone (PTH) and PTH-related protein analogs in transmembrane domain two of the type 1 PTH receptor. _Mol. Endocrinol._ 17, 2647–2658 (2003) Article CAS PubMed
Google Scholar * de Graaf, C., Rein, C., Piwnica, D., Giordanetto, F. & Rognan, D. Structure-based discovery of allosteric modulators of two related class B G-protein-coupled receptors.
_ChemMedChem_ 6, 2159–2169 (2011) Article CAS PubMed Google Scholar * Chun, E. et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors.
_Structure_ 20, 967–976 (2012) Article CAS PubMed PubMed Central Google Scholar * Ballesteros, J. A. & Weinstein, H. in _Methods in Neurosciences_ Vol. 25 (ed. Sealfon, S. C. )
366–428 (Academic Press, 1995) Google Scholar * Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of
ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. _Proc. Natl Acad. Sci. USA_ 110, 5211–5216 (2013) Article CAS ADS PubMed PubMed Central
Google Scholar * Unson, C. G. et al. Roles of specific extracellular domains of the glucagon receptor in ligand binding and signaling. _Biochemistry_ 41, 11795–11803 (2002) Article CAS
PubMed Google Scholar * Xiao, Q., Jeng, W. & Wheeler, M. B. Characterization of glucagon-like peptide-1 receptor-binding determinants. _J. Mol. Endocrinol._ 25, 321–335 (2000) Article
CAS PubMed Google Scholar * Roberts, D. J., Vertongen, P. & Waelbroeck, M. Analysis of the glucagon receptor first extracellular loop by the substituted cysteine accessibility
method. _Peptides_ 32, 1593–1599 (2011) Article CAS PubMed Google Scholar * Wu, H. et al. Structure of the human κ-opioid receptor in complex with JDTic. _Nature_ 485, 327–332 (2012)
Article CAS ADS PubMed PubMed Central Google Scholar * Manglik, A. et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. _Nature_ 485, 321–326 (2012)
Article CAS ADS PubMed PubMed Central Google Scholar * Fredriksson, R., Lagerstrom, M. C., Lundin, L. G. & Schioth, H. B. The G-protein-coupled receptors in the human genome form
five main families. Phylogenetic analysis, paralogon groups, and fingerprints. _Mol. Pharmacol._ 63, 1256–1272 (2003) Article CAS PubMed Google Scholar * Venkatakrishnan, A. J. et al.
Molecular signatures of G-protein-coupled receptors. _Nature_ 494, 185–194 (2013) Article CAS ADS PubMed Google Scholar * Inooka, H. et al. Conformation of a peptide ligand bound to its
G-protein coupled receptor. _Nature Struct. Biol._ 8, 161–165 (2001) Article CAS PubMed Google Scholar * Prévost, M. et al. Mutational and cysteine scanning analysis of the glucagon
receptor N-terminal domain. _J. Biol. Chem._ 285, 30951–30958 (2010) Article PubMed PubMed Central Google Scholar * Ahn, J. M., Medeiros, M., Trivedi, D. & Hruby, V. J. Development
of potent truncated glucagon antagonists. _J. Med. Chem._ 44, 1372–1379 (2001) Article CAS PubMed Google Scholar * Coopman, K. et al. Residues within the transmembrane domain of the
glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor. _Mol. Endocrinol._ 25, 1804–1818 (2011) Article CAS PubMed PubMed
Central Google Scholar * Di Paolo, E. et al. Contribution of the second transmembrane helix of the secretin receptor to the positioning of secretin. _FEBS Lett._ 424, 207–210 (1998)
Article CAS PubMed Google Scholar * Perret, J. et al. Mutational analysis of the glucagon receptor: similarities with the vasoactive intestinal peptide (VIP)/pituitary adenylate
cyclase-activating peptide (PACAP)/secretin receptors for recognition of the ligand's third residue. _Biochem. J._ 362, 389–394 (2002) CAS PubMed PubMed Central Google Scholar *
Runge, S. et al. Three distinct epitopes on the extracellular face of the glucagon receptor determine specificity for the glucagon amino terminus. _J. Biol. Chem._ 278, 28005–28010 (2003)
Article CAS PubMed Google Scholar * Neumann, J. M. et al. Class-B GPCR activation: is ligand helix-capping the key? _Trends Biochem. Sci._ 33, 314–319 (2008) Article CAS PubMed Google
Scholar * Cascieri, M. A. et al. Characterization of a novel, non-peptidyl antagonist of the human glucagon receptor. _J. Biol. Chem._ 274, 8694–8697 (1999) Article CAS PubMed Google
Scholar * Di Paolo, E. et al. Mutations of aromatic residues in the first transmembrane helix impair signalling by the secretin receptor. _Receptors Channels_ 6, 309–315 (1999) CAS PubMed
Google Scholar * Koole, C. et al. Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation. _J.
Biol. Chem._ 287, 3642–3658 (2012) Article CAS PubMed Google Scholar * Tseng, C. C. & Lin, L. A point mutation in the glucose-dependent insulinotropic peptide receptor confers
constitutive activity. _Biochem. Biophys. Res. Commun._ 232, 96–100 (1997) Article CAS PubMed Google Scholar * Ganguli, S. C. et al. Protean effects of a natural peptide agonist of the G
protein-coupled secretin receptor demonstrated by receptor mutagenesis. _J. Pharmacol. Exp. Ther._ 286, 593–598 (1998) CAS PubMed Google Scholar * Solano, R. M. et al. Two basic residues
of the h-VPAC1 receptor second transmembrane helix are essential for ligand binding and signal transduction. _J. Biol. Chem._ 276, 1084–1088 (2001) Article CAS PubMed Google Scholar *
Ceraudo, E. et al. Spatial proximity between the VPAC1 receptor and the amino terminus of agonist and antagonist peptides reveals distinct sites of interaction. _FASEB J._ 26, 2060–2071
(2012) Article CAS PubMed Google Scholar * Tan, Y. V., Couvineau, A. & Laburthe, M. Diffuse pharmacophoric domains of vasoactive intestinal peptide (VIP) and further insights into
the interaction of VIP with the N-terminal ectodomain of human VPAC1 receptor by photoaffinity labeling with [Bpa6]-VIP. _J. Biol. Chem._ 279, 38889–38894 (2004) Article CAS PubMed Google
Scholar * Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. _Nature Protocols_ 4, 706–731 (2009) Article CAS PubMed PubMed Central Google Scholar
* Di Paolo, E. et al. Role of charged amino acids conserved in the vasoactive intestinal polypeptide/secretin family of receptors on the secretin receptor functionality. _Peptides_ 20,
1187–1193 (1999) Article CAS PubMed Google Scholar * Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for
positioning of proteins in membranes. _Nucleic Acids Res._ 40, D370–D376 (2012) Article CAS PubMed Google Scholar * Hanson, M. A. et al. Profiling of membrane protein variants in a
baculovirus system by coupling cell-surface detection with small-scale parallel expression. _Protein Expr. Purif._ 56, 85–92 (2007) Article CAS PubMed PubMed Central Google Scholar *
Misquitta, Y. & Caffrey, M. Detergents destabilize the cubic phase of monoolein: implications for membrane protein crystallization. _Biophys. J._ 85, 3084–3096 (2003) Article CAS ADS
PubMed PubMed Central Google Scholar * Cherezov, V., Peddi, A., Muthusubramaniam, L., Zheng, Y. F. & Caffrey, M. A robotic system for crystallizing membrane and soluble proteins in
lipidic mesophases. _Acta Crystallogr. D_ 60, 1795–1807 (2004) Article PubMed Google Scholar * Xu, F., Liu, W., Hanson, M. A., Stevens, R. C. & Cherezov, V. Development of an
automated high throughput LCP-FRAP assay to guide membrane protein crystallization in lipid mesophases. _Cryst. Growth Des._ 11, 1193–1201 (2011) Article CAS PubMed PubMed Central Google
Scholar * Cherezov, V. et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 µm size X-ray synchrotron beam. _J. R. Soc.
Interface_ 6 (Suppl 5). S587–S597 (2009) Article CAS PubMed PubMed Central Google Scholar * Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation
mode. _Methods Enzymol._ 276, 307–326 (1997) Article CAS PubMed Google Scholar * McCoy, A. J. et al. Phaser crystallographic software. _J. Appl. Crystalogr._ 40, 658–674 (2007) Article
CAS Google Scholar * Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the _PHENIX AutoSol_ wizard. _Acta Crystallogr. D_ 65, 582–601
(2009) Article CAS PubMed PubMed Central Google Scholar * Terwilliger, T. C. et al. _phenix._ _mr_rosetta_: molecular replacement and model rebuilding with _Phenix_ and _Rosetta_. _J.
Struct. Funct. Genomics_ 13, 81–90 (2012) Article CAS PubMed PubMed Central Google Scholar * Schwarzenbacher, R., Godzik, A. & Jaroszewski, L. The JCSG MR pipeline: optimized
alignments, multiple models and parallel searches. _Acta Crystallogr. D_ 64, 133–140 (2008) Article CAS PubMed Google Scholar * Murshudov, G. N., Vagin, A. A. & Dodson, E. J.
Refinement of macromolecular structures by the maximum-likelihood method. _Acta Crystallogr. D_ 53, 240–255 (1997) Article CAS PubMed Google Scholar * BUSTER. v. 2.8.0 (Global Phasing,
Cambridge, UK, 2009) * Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the _PHENIX AutoBuild_ wizard. _Acta Crystallogr. D_ 64, 61–69
(2008) Article CAS PubMed Google Scholar * Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of _Coot_. _Acta Crystallogr. D_ 66, 486–501 (2010) Article
CAS PubMed PubMed Central Google Scholar * Bhat, T. Calculation of an OMIT map. _J. Appl. Crystallogr._ 21, 279–281 (1988) Article Google Scholar * ICM. Manual v. 3.0 (MolSoft, La
Jolla, California, 2012) * Arnautova, Y. A., Abagyan, R. A. & Totrov, M. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop
modeling. _Proteins_ 79, 477–498 (2011) Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by NIH Roadmap grant P50 GM073197
for technology development (V.C. and R.C.S.), and PSI:Biology grant U54 GM094618 for biological studies and structure production (target GPCR-49) (V.K., V.C. and R.C.S.); PSI:Biology grant
U54 GM094586 for structure QC; The Ministry of Health grants 2012ZX09304-011 and 2013ZX09507002 (M.-W.W.), Shanghai Science and Technology Development Fund 11DZ2292200 (M.-W.W.); Novo
Nordisk-Chinese Academy of Sciences Research Fund NNCAS-2011-7 (M.-W.W.); Thousand Talents Program in China (R.C.S. and M.-W.W.); NIH Postdoctoral Training Grant (NRSA) F32 DK088392
(F.Y.S.); The Netherlands Organization for Scientific Research (NWO) through a VENI grant (Grant 700.59.408 to C.d.G.); COST Action CM1207, GLISTEN (C.d.G). We also thank V. Hruby and M. Cai
for advice with the glucagon binding assay and general discussions; J. Velasquez for help with molecular biology; T. Trinh and M. Chu for help with baculovirus expression; K. Kadyshevskaya
for assistance with figure preparation; X. Q. Cai, J. Wang, Y. Feng, A. T. Dai, Y. Zhou, J. J. Deng, Y. B. Dai and J. W. Zhao for technical assistance in mutation studies; A. Walker for
assistance with manuscript preparation; and J. Smith and R. Fischetti for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon
Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS *
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, USA, Fai Yiu Siu, Gye Won Han,
Daniel Wacker, Jeremiah S. Joseph, Wei Liu, Vadim Cherezov, Vsevolod Katritch & Raymond C. Stevens * The National Center for Drug Screening and the CAS Key Laboratory of Receptor
Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (CAS), 189 Guo Shou Jing Road, Shanghai, 201203, China, Min He, Dehua Yang, Zhiyun Zhang, Caihong Zhou &
Ming-Wei Wang * Division of Medicinal Chemistry, Faculty of Sciences, Amsterdam Institute for Molecules, Medicines and Systems (AIMMS), VU University of Amsterdam, De Boelelaan 1083, 1081 HV
Amsterdam, The Netherlands, Chris de Graaf * The Joint Center for Structural Genomics, Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park,
California 94025, USA, Qingping Xu * Protein & Peptide Chemistry, Novo Nordisk, Novo Nordisk Park, 2760 Malov, Denmark, Jesper Lau Authors * Fai Yiu Siu View author publications You can
also search for this author inPubMed Google Scholar * Min He View author publications You can also search for this author inPubMed Google Scholar * Chris de Graaf View author publications
You can also search for this author inPubMed Google Scholar * Gye Won Han View author publications You can also search for this author inPubMed Google Scholar * Dehua Yang View author
publications You can also search for this author inPubMed Google Scholar * Zhiyun Zhang View author publications You can also search for this author inPubMed Google Scholar * Caihong Zhou
View author publications You can also search for this author inPubMed Google Scholar * Qingping Xu View author publications You can also search for this author inPubMed Google Scholar *
Daniel Wacker View author publications You can also search for this author inPubMed Google Scholar * Jeremiah S. Joseph View author publications You can also search for this author inPubMed
Google Scholar * Wei Liu View author publications You can also search for this author inPubMed Google Scholar * Jesper Lau View author publications You can also search for this author
inPubMed Google Scholar * Vadim Cherezov View author publications You can also search for this author inPubMed Google Scholar * Vsevolod Katritch View author publications You can also search
for this author inPubMed Google Scholar * Ming-Wei Wang View author publications You can also search for this author inPubMed Google Scholar * Raymond C. Stevens View author publications
You can also search for this author inPubMed Google Scholar CONTRIBUTIONS F.Y.S. designed, expressed, characterized and screened constructs and ligands for crystallization. F.Y.S. purified
and crystallized the receptor in LCP, optimized crystallization conditions, grew crystals, collected diffraction data and prepared the manuscript. G.W.H. and Q.X. solved and refined the
structure, and prepared the manuscript. V.C. collected and processed diffraction data, and prepared the manuscript. M.H., D.Y., Z.Z. and C.Z. expressed the receptor, and performed the
mutagenesis and ligand-binding assay. V.K. and C.d.G. designed and analysed the receptor mutagenesis studies, constructed the receptor–ligand model and prepared the manuscript. D.W. and
J.S.J. collected and processed SAD data and determined an initial electron density map from experimental phases. W.L. and V.C. trained and assisted in LCP crystallization. J.L. provided
ligands for GCGR and prepared the manuscript. R.C.S., F.Y.S., M.-W.W., V.K., V.C. and C.d.G. were responsible for the overall project strategy and management and wrote the manuscript.
CORRESPONDING AUTHORS Correspondence to Ming-Wei Wang or Raymond C. Stevens. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION This file contains Supplementary Tables 1-6, Supplementary Figures 1-10 and Supplementary References. (PDF 1596 kb) POWERPOINT SLIDES POWERPOINT SLIDE
FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 POWERPOINT SLIDE FOR FIG. 5 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Siu, F., He, M., de Graaf, C. _et al._ Structure of the human glucagon class B G-protein-coupled receptor. _Nature_ 499, 444–449 (2013). https://doi.org/10.1038/nature12393
Download citation * Received: 07 March 2013 * Accepted: 17 June 2013 * Published: 17 July 2013 * Issue Date: 25 July 2013 * DOI: https://doi.org/10.1038/nature12393 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative


