- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
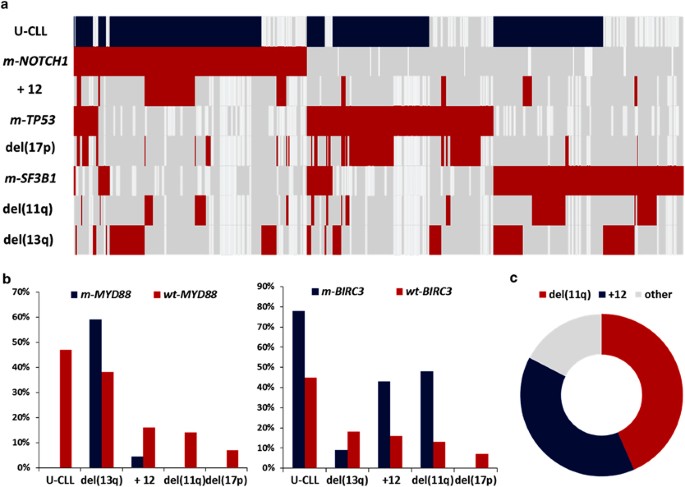
ABSTRACT Through the European Research Initiative on chronic lymphocytic leukemia (CLL) (ERIC), we screened 3490 patients with CLL for mutations within the _NOTCH1_ (_n_=3334), _SF3B1_
(_n_=2322), _TP53_ (_n_=2309), _MYD88_ (_n_=1080) and _BIRC3_ (_n_=919) genes, mainly at diagnosis (75%) and before treatment (>90%). _BIRC3_ mutations (2.5%) were associated with
unmutated _IGHV_ genes (U-CLL), del(11q) and trisomy 12, whereas _MYD88_ mutations (2.2%) were exclusively found among M-CLL. _NOTCH1_, _SF3B1_ and _TP53_ exhibited variable frequencies and
were mostly enriched within clinically aggressive cases. Interestingly, as the timespan between diagnosis and mutational screening increased, so too did the incidence of _SF3B1_ mutations;
no such increase was observed for _NOTCH1_ mutations. Regarding the clinical impact, _NOTCH1_ mutations, _SF3B1_ mutations and _TP53_ aberrations (deletion/mutation, _TP53ab_) correlated
with shorter time-to-first-treatment (_P_<0.0001) in 889 treatment-naive Binet stage A cases. In multivariate analysis (_n_=774), _SF3B1_ mutations and _TP53ab_ along with del(11q) and
U-CLL, but not _NOTCH1_ mutations, retained independent significance. Importantly, _TP53ab_ and _SF3B1_ mutations had an adverse impact even in U-CLL. In conclusion, we support the clinical
relevance of novel recurrent mutations in CLL, highlighting the adverse impact of _SF3B1_ and _TP53_ mutations, even independent of _IGHV_ mutational status, thus underscoring the need for
urgent standardization/harmonization of the detection methods. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS DIFFERENT PROGNOSTIC IMPACT OF RECURRENT GENE MUTATIONS IN CHRONIC LYMPHOCYTIC
LEUKEMIA DEPENDING ON IGHV GENE SOMATIC HYPERMUTATION STATUS: A STUDY BY ERIC IN HARMONY Article Open access 24 December 2022 _ATM_ ABERRATIONS IN CHRONIC LYMPHOCYTIC LEUKEMIA: DEL(11Q)
RATHER THAN _ATM_ MUTATIONS IS AN ADVERSE-PROGNOSTIC BIOMARKER Article Open access 24 April 2025 TIME TO FIRST TREATMENT AND P53 DYSFUNCTION IN CHRONIC LYMPHOCYTIC LEUKAEMIA: RESULTS OF THE
O-CLL1 STUDY IN EARLY STAGE PATIENTS Article Open access 28 October 2020 REFERENCES * Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J _et al_. Analysis of the chronic lymphocytic
leukemia coding genome: role of NOTCH1 mutational activation. _J Exp Med_ 2011; 208: 1389–1401. Article CAS PubMed PubMed Central Google Scholar * Puente XS, Pinyol M, Quesada V, Conde
L, Ordonez GR, Villamor N _et al_. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. _Nature_ 2011; 475: 101–105. Article CAS PubMed PubMed Central
Google Scholar * Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L _et al_. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic
lymphocytic leukemia. _Nat Genet_ 2012; 44: 47–52. Article CAS Google Scholar * Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K _et al_. SF3B1 and other novel cancer genes
in chronic lymphocytic leukemia. _N Engl J Med_ 2011; 365: 2497–2506. Article CAS PubMed PubMed Central Google Scholar * Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner
K _et al_. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. _Blood_ 2014; 123: 3247–3254. Article CAS PubMed Google Scholar * Schnaiter
A, Paschka P, Rossi M, Zenz T, Buhler A, Winkler D _et al_. NOTCH1, SF3B1, and TP53 mutations in fludarabine-refractory CLL patients treated with alemtuzumab: results from the CLL2H trial of
the GCLLSG. _Blood_ 2013; 122: 1266–1270. Article CAS PubMed Google Scholar * Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M _et al_. Mutations of the SF3B1 splicing
factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. _Blood_ 2011; 118: 6904–6908. Article CAS PubMed PubMed Central Google Scholar *
Cortese D, Sutton LA, Cahill N, Smedby KE, Geisler C, Gunnarsson R _et al_. On the way towards a 'CLL prognostic index': focus on TP53, BIRC3, SF3B1, NOTCH1 and MYD88 in a
population-based cohort. _Leukemia_ 2013; 28: 710–713. Article PubMed Google Scholar * Del Giudice I, Rossi D, Chiaretti S, Marinelli M, Tavolaro S, Gabrielli S _et al_. NOTCH1 mutations
in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. _Haematologica_
2012; 97: 437–441. Article CAS PubMed PubMed Central Google Scholar * Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V _et al_. SF3B1 mutations correlated to
cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. _Leukemia_ 2014; 28: 108–117. Article CAS PubMed Google Scholar * Oscier DG,
Rose-Zerilli MJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J _et al_. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. _Blood_ 2013; 121: 468–475.
Article CAS PubMed Google Scholar * Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, Cresta S _et al_. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53
wild-type chronic lymphocytic leukemia. _Blood_ 2012; 119: 2854–2862. Article CAS PubMed Google Scholar * Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C _et al_. Integrated
mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. _Blood_ 2013; 121: 1403–1412. Article CAS PubMed PubMed Central Google Scholar
* Weissmann S, Roller A, Jeromin S, Hernandez M, Abaigar M, Hernandez-Rivas JM _et al_. Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on
852 patients. _Leukemia_ 2013; 27: 2393–2396. Article CAS PubMed Google Scholar * Villamor N, Conde L, Martinez-Trillos A, Cazorla M, Navarro A, Bea S _et al_. NOTCH1 mutations identify
a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. _Leukemia_ 2013; 27: 1100–1106. Article CAS PubMed Google Scholar *
Mansouri L, Cahill N, Gunnarsson R, Smedby KE, Tjonnfjord E, Hjalgrim H _et al_. NOTCH1 and SF3B1 mutations can be added to the hierarchical prognostic classification in chronic lymphocytic
leukemia. _Leukemia_ 2013; 27: 512–514. Article CAS PubMed Google Scholar * Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R _et al_. Monitoring chronic lymphocytic leukemia
progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. _Blood_ 2012; 120: 4191–4196. Article CAS PubMed Google Scholar * Schwaederle M, Ghia E, Rassenti
LZ, Obara M, Dell'Aquila ML, Fecteau JF _et al_. Subclonal evolution involving SF3B1 mutations in chronic lymphocytic leukemia. _Leukemia_ 2013; 27: 1214–1217. Article CAS PubMed
PubMed Central Google Scholar * Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS _et al_. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia.
_Cell_ 2013; 152: 714–726. Article CAS PubMed PubMed Central Google Scholar * Trbusek M, Smardova J, Malcikova J, Sebejova L, Dobes P, Svitakova M _et al_. Missense mutations located in
structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. _J Clin Oncol_ 2011; 29: 2703–2708. Article CAS PubMed Google Scholar *
Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ _et al_. NOTCH1 mutations in CLL associated with trisomy 12. _Blood_ 2012; 119: 329–331. Article CAS PubMed PubMed
Central Google Scholar * Lopez C, Delgado J, Costa D, Conde L, Ghita G, Villamor N _et al_. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy
12 or associated with other chromosomal alterations. _Genes Chromosomes Cancer_ 2012; 51: 881–889. Article CAS PubMed Google Scholar * Dohner H, Stilgenbauer S, Benner A, Leupolt E,
Krober A, Bullinger L _et al_. Genomic aberrations and survival in chronic lymphocytic leukemia. _N Engl J Med_ 2000; 343: 1910–1916. Article CAS PubMed Google Scholar * Gunnarsson R,
Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N _et al_. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic
leukemia: a high-resolution genomic screening of newly diagnosed patients. _Leukemia_ 2010; 24: 211–215. Article CAS PubMed Google Scholar * Agathangelidis A, Darzentas N, Hadzidimitriou
A, Brochet X, Murray F, Yan XJ _et al_. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies.
_Blood_ 2012; 119: 4467–4475. Article CAS PubMed PubMed Central Google Scholar * Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C _et al_. Stereotyped patterns
of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. _Blood_ 2008; 111: 1524–1533. Article
CAS PubMed Google Scholar * Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H _et al_. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia:
a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. _Blood_ 2008; 111: 5446–5456. Article CAS
PubMed PubMed Central Google Scholar * Xu L, Hunter ZR, Yang G, Zhou Y, Cao Y, Liu X _et al_. MYD88 L265P in Waldenstrom macroglobulinemia, immunoglobulin M monoclonal gammopathy, and
other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. _Blood_ 2013; 121: 2051–2058. Article CAS PubMed PubMed Central
Google Scholar * Rose-Zerilli MJ, Forster J, Parker H, Parker A, Rodriguez AE, Chaplin T _et al_. ATM mutation rather than BIRC3 deletion and/or mutation predicts reduced survival in
11q-deleted chronic lymphocytic leukemia: data from the UK LRF CLL4 trial. _Haematologica_ 2014; 99: 736–742. Article CAS PubMed PubMed Central Google Scholar * Shedden K, Li Y,
Ouillette P, Malek SN . Characteristics of chronic lymphocytic leukemia with somatically acquired mutations in NOTCH1 exon 34. _Leukemia_ 2012; 26: 1108–1110. Article CAS PubMed Google
Scholar * Gentien D, Kosmider O, Nguyen-Khac F, Albaud B, Rapinat A, Dumont AG _et al_. A common alternative splicing signature is associated with SF3B1 mutations in malignancies from
different cell lineages. _Leukemia_ 2014; 28: 1355–1357. Article CAS PubMed Google Scholar * Rahal R, Frick M, Romero R, Korn JM, Kridel R, Chan FC _et al_. Pharmacological and genomic
profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma. _Nat Med_ 2014; 20: 87–92. Article CAS PubMed Google Scholar * Oscier DG, Gardiner AC, Mould SJ,
Glide S, Davis ZA, Ibbotson RE _et al_. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent
prognostic factors. _Blood_ 2002; 100: 1177–1184. CAS PubMed Google Scholar * Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P _et al_. V(H) mutation status, CD38
expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. _Blood_ 2002; 100: 1410–1416. CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We are
grateful to the European Research Initiative in CLL steering board for all their support during this project. We thank Jitka Malcikova and Karla Plevova for their valuable comments and help
in sample management and to Jana Smardova for FASAY analysis. This research was funded by the Nordic Cancer Union, the Swedish Cancer Society, the Swedish Research Council and Lion’s Cancer
Research Foundation, Uppsala; Cancer Research UK, Leukaemia and Lymphoma Research, Kay Kendall Leukaemia Fund, United Kingdom; Associazione Italiana per la Ricerca sul Cancro (AIRC)
(Investigator grant and Molecular Clinical Oncology Program 5xMille no. 9965, no. 10007), Milano, Italy; Ricerca Finalizzata 2010, Ministero della Salute, Roma, Italy; the ENosAI project
(code 09SYN-13-880) co-funded by the EU and the Hellenic General Secretariat for Research and Technology, Greece; by project MSMT-CR VaVPI CZ.1.05/1.1.00/02.0068 (CEITEC), research grant
IGA-MZ-CR NT13493-4/2012 and 7th FP Health project NGS-PTL/2012-2015/no. 306242 of European Commission and MSMT-CR (2013-2015, no. 7E13008), and the ICGC-CLL Genome Project from the Spanish
Ministry of Economy and Competitivity (MINECO) through the Instituto de Salud Carlos III (RTICC RD12/0036/0023 and PIE PI043043), Spain. Andreas Agathangelidis is a recipient of a fellowship
by Associazione Italiana per la Ricerca sul Cancro AIRC (Triennial fellowship ‘Guglielmina Lucatello é Gino Mazzega’). AUTHOR INFORMATION Author notes * J C Strefford, K Stamatopoulos and R
Rosenquist: The last three authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Immunology, Science for Life Laboratory, Genetics and Pathology, Rudbeck
Laboratory, Uppsala University, Uppsala, Sweden P Baliakas, A Hadzidimitriou, L-A Sutton, L Mansouri, D Cortese, K Stamatopoulos & R Rosenquist * Hematology Department and HCT Unit, G
Papanicolaou Hospital, Thessaloniki, Greece P Baliakas, E Stalika, A Anagnostopoulos & K Stamatopoulos * Institute of Applied Biosciences, CERTH, Thessaloniki, Greece A Hadzidimitriou, E
Minga, A M Makris & K Stamatopoulos * Division of Haematology, Department of Translational Medicine, Amedeo Avogadro University of Eastern Piedmont, Novara, Italy D Rossi & G
Gaidano * Hematopathology Unit and Department of Hematology, Hospital Clinic, University of Barcelona, Institut d’Investigacions Biomèdiques August Pi iSunyer (IDIBAPS), Barcelona, Spain N
Villamor, A Navarro, J Delgado & E Campo * Cancer Sciences, Faculty of Medicine, University of Southampton, Southampton, UK M Larrayoz, M J J Rose-Zerilli & J C Strefford * Central
European Institute of Technology, Masaryk University and University Hospital Brno, Brno, Czech Republic J Kminkova, B Kantorova, V Navrkalova & S Pospisilova * Università Vita-Salute San
Raffaele, Milan, Italy A Agathangelidis, L Scarfò & P Ghia * Division of Molecular Oncology and Department of Onco-Hematology, San Raffaele Scientific Institute, Milan, Italy A
Agathangelidis, L Scarfò & P Ghia * Department of Haematology, Royal Bournemouth Hospital, Bournemouth, UK Z Davis & D Oscier * Department of Internal Medicine III, Ulm University,
Ulm, Germany E Tausch & S Stilgenbauer * Department of Medicine, Solna, Clinical Epidemiology Unit, Karolinska Institutet, Stockholm, Sweden K E Smedby * Lund University and Hospital
Department of Hematology, Lund Stem Cell Center, Lund, Sweden G Juliusson * Hematology Department, Nikea General Hospital, Pireaus, Greece C Belessi Authors * P Baliakas View author
publications You can also search for this author inPubMed Google Scholar * A Hadzidimitriou View author publications You can also search for this author inPubMed Google Scholar * L-A Sutton
View author publications You can also search for this author inPubMed Google Scholar * D Rossi View author publications You can also search for this author inPubMed Google Scholar * E Minga
View author publications You can also search for this author inPubMed Google Scholar * N Villamor View author publications You can also search for this author inPubMed Google Scholar * M
Larrayoz View author publications You can also search for this author inPubMed Google Scholar * J Kminkova View author publications You can also search for this author inPubMed Google
Scholar * A Agathangelidis View author publications You can also search for this author inPubMed Google Scholar * Z Davis View author publications You can also search for this author
inPubMed Google Scholar * E Tausch View author publications You can also search for this author inPubMed Google Scholar * E Stalika View author publications You can also search for this
author inPubMed Google Scholar * B Kantorova View author publications You can also search for this author inPubMed Google Scholar * L Mansouri View author publications You can also search
for this author inPubMed Google Scholar * L Scarfò View author publications You can also search for this author inPubMed Google Scholar * D Cortese View author publications You can also
search for this author inPubMed Google Scholar * V Navrkalova View author publications You can also search for this author inPubMed Google Scholar * M J J Rose-Zerilli View author
publications You can also search for this author inPubMed Google Scholar * K E Smedby View author publications You can also search for this author inPubMed Google Scholar * G Juliusson View
author publications You can also search for this author inPubMed Google Scholar * A Anagnostopoulos View author publications You can also search for this author inPubMed Google Scholar * A M
Makris View author publications You can also search for this author inPubMed Google Scholar * A Navarro View author publications You can also search for this author inPubMed Google Scholar
* J Delgado View author publications You can also search for this author inPubMed Google Scholar * D Oscier View author publications You can also search for this author inPubMed Google
Scholar * C Belessi View author publications You can also search for this author inPubMed Google Scholar * S Stilgenbauer View author publications You can also search for this author
inPubMed Google Scholar * P Ghia View author publications You can also search for this author inPubMed Google Scholar * S Pospisilova View author publications You can also search for this
author inPubMed Google Scholar * G Gaidano View author publications You can also search for this author inPubMed Google Scholar * E Campo View author publications You can also search for
this author inPubMed Google Scholar * J C Strefford View author publications You can also search for this author inPubMed Google Scholar * K Stamatopoulos View author publications You can
also search for this author inPubMed Google Scholar * R Rosenquist View author publications You can also search for this author inPubMed Google Scholar CONSORTIA ON BEHALF OF THE EUROPEAN
RESEARCH INITIATIVE ON CLL (ERIC) CORRESPONDING AUTHOR Correspondence to R Rosenquist. ETHICS DECLARATIONS COMPETING INTERESTS KS receives research support from Roche SA. JCS has financial
support from Hoffman La Roche. ADDITIONAL INFORMATION Supplementary Information accompanies this paper on the Leukemia website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 565
KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Baliakas, P., Hadzidimitriou, A., Sutton, LA. _et al._ Recurrent mutations refine prognosis in
chronic lymphocytic leukemia. _Leukemia_ 29, 329–336 (2015). https://doi.org/10.1038/leu.2014.196 Download citation * Received: 09 April 2014 * Revised: 20 May 2014 * Accepted: 11 June 2014
* Published: 19 June 2014 * Issue Date: February 2015 * DOI: https://doi.org/10.1038/leu.2014.196 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative

![[withdrawn] near miss with a track worker at llandegai tunnel](https://assets.publishing.service.gov.uk/media/6051d710d3bf7f0455a6e604/s960_Llandegai_tunnel.jpg)





