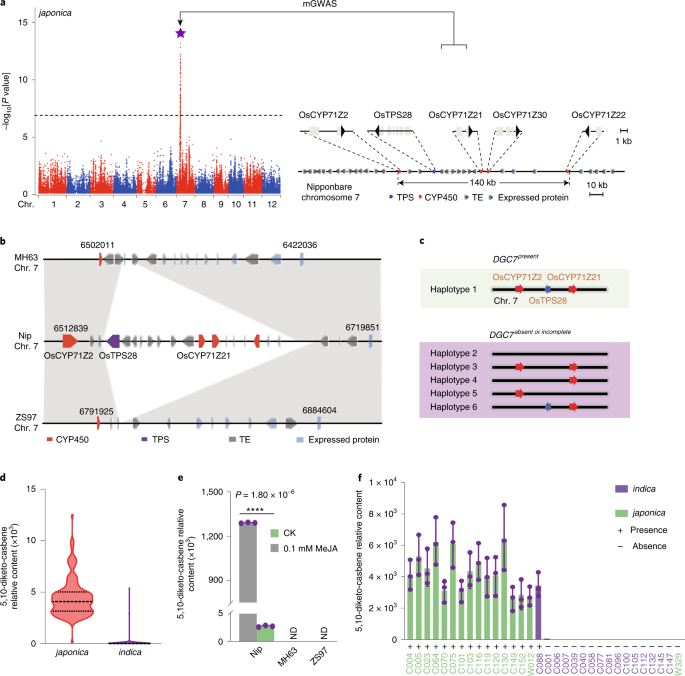
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Small cell lung cancer (SCLC) constitutes approximately 15% of all diagnosed lung cancers. SCLC is a particularly lethal malignancy, as the 2-year survival rate after appropriate
treatment is less than 5%. The patients with SCLC have not been received a benefit of the recently developed molecular targeted treatment. Therefore, a new treatment strategy is necessary
for the patients. The molecular mechanisms underlying the aggressiveness of SCLC cells and their development of treatment-resistance are still ambiguous. In this study, we newly constructed
a microRNA (miRNA) expression signature of SCLC by analysis of autopsy specimens. Based on the resultant signature, four miRNAs (_miR-27a-5p_, _miR-485-3p_, _miR-34-5p_ and _miR-574-3p_)
were found to be candidate anti-tumor miRNAs. To investigate their functional importance, we first validated the downregulation of _miR-27a-5p_ and _miR-34b-3p_ in SCLC clinical specimens.
Next, we demonstrated that ectopic expression of both _miR-27a-5p_ and _miR-34b-3p_ significantly inhibited cancer cell aggressiveness. Our _in silico_ analyses showed that four genes
(topoisomerase 2 alpha (_TOP2A_), maternal embryonic leucine zipper kinase (_MELK_), centromere protein F (_CENPF_) and SRY-box 1 (_SOX1_) were identified as _miR-27a-5p-_ and
_miR-34b-3p_-regulated genes. Based on immunohistochemical analysis, TOP2A, MELK and CENPF were involved in SCLC pathogenesis. These genes might contribute to high proliferation and early
metastatic spread of SCLC cells. Elucidation of differentially expressed miRNA-mediated cancer pathways based on SCLC signature may provide new insights into the mechanisms of SCLC
pathogenesis. SIMILAR CONTENT BEING VIEWED BY OTHERS MIR-494-3P ENHANCES AGGRESSIVE PHENOTYPE OF NON-SMALL CELL LUNG CANCER CELLS BY REGULATING SET/I2PP2A Article Open access 02 May 2025
MICRORNA-150-3P ENHANCES THE ANTITUMOUR EFFECTS OF CGP57380 AND IS ASSOCIATED WITH A FAVOURABLE PROGNOSIS IN NON-SMALL CELL LUNG CANCER Article Open access 15 January 2025 MIR-4634 AUGMENTS
THE ANTI-TUMOR EFFECTS OF RAD001 AND ASSOCIATES WELL WITH CLINICAL PROGNOSIS OF NON-SMALL CELL LUNG CANCER Article Open access 04 August 2020 INTRODUCTION Small cell lung cancer (SCLC)
constitutes approximately 15–20% of all diagnosed lung cancers.1, 2 Owing to its aggressive nature, SCLC is a particularly lethal malignancy. SCLC cells characteristically acquire rapid cell
proliferation and the ability to metastasize to distant sites. The majority of patients with SCLC present with metastatic disease, and the median survival time with combination chemotherapy
is under 1 year for patients with extensive disease.1, 2 The conventional first-line treatment for SCLC with extensive disease is platinum-based chemotherapy.3, 4, 5 Although the response
rate of the treatment is good (70–80%), cancer cells acquire early resistance to conventional treatments. Median progression-free survival is 5–6 months,3, 4, 5 and the disease escalates its
aggressiveness. The patients with SCLC have not been received a benefit of the recently developed molecular targeted treatment. Therefore, understanding the molecular mechanisms of SCLC
aggressiveness through current genomic approaches is needed. The discovery of non-coding RNA in the human genome provided new directions for the study of human cancer pathogenesis.6
MicroRNAs (miRNAs) belong to a member of non-coding RNAs that act as sequence-specific fine tuners of the expression levels of proteins and RNAs.7, 8 A single miRNA can regulate a large
number of RNA transcripts in human cells.9 Thus, aberrantly expressed miRNAs cause disruption of tightly regulated RNA networks, leading to pathologic behavior of cancer cells.10, 11
Currently, numerous studies have indicated that aberrantly expressed miRNAs are deeply involved in cancer pathogenesis.10, 11 We have been revealed the anti-tumor miRNAs and their controlled
cancer pathways by using miRNA expression signatures of several cancers, including lung cancer.12, 13, 14 The next challenge in our miRNA studies is to identify key molecules and novel
pathways involved in the resistance of cancer cells to current treatments. Based on this, we have constructed miRNA expression signatures by analyzing autopsy specimens from patients with
prostate cancer and renal cell carcinoma.15, 16 Based on these signatures, we previously identified tumor-suppressive _miR-221/222_-mediated castration-resistant prostate cancer pathways and
_miR-101_-mediated sunitinib-resistant pathways.15, 16 The molecular mechanisms underlying the aggressiveness of SCLC cells and the development of therapy-resistance are still ambiguous.
Thus, we have investigated the molecular pathways contributing to treatment-resistant cancer cells in an effort to develop the new therapeutic strategies in SCLC. In the present study, we
newly constructed miRNA expression signatures through analysis of primary and metastatic lesions (liver and brain). Based on the signatures, we identified two tumor-suppressive miRNAs
(_miR-27a-5p_ and _miR-34b-3p_) that are deeply involved in SCLC pathogenesis. Elucidation of the miRNA signature of SCLC may be useful for identification of novel molecular mechanisms of
SCLC recurrence, metastasis and drug resistance. MATERIALS AND METHODS PATIENTS AND CLINICAL LUNG CANCER SPECIMENS Clinical lung specimens were obtained from patients admitted to the
Kagoshima University Hospital from 2011 to 2015. The patients’ backgrounds and clinical characteristics are summarized in Table 1 and Supplementary Table S1. Normal tissues are summarized in
Supplementary Table S2. Archival formalin-fixed, paraffin-embedded samples were used for expression analysis and immunohistochemistry. Clinical specimens were staged according to the
International Association for the Study of Lung Cancer TNM classification.17 This protocol was approved by the Institutional Review Board for Clinical Research of Kagoshima University School
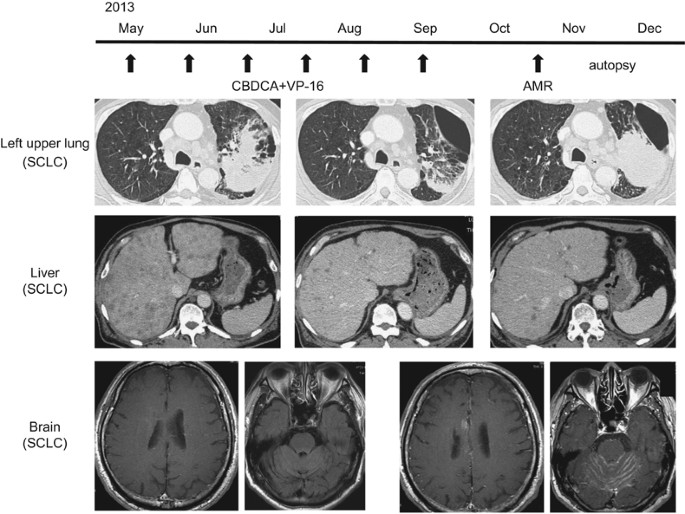
of Medicine. CONSTRUCTION OF THE MIRNA EXPRESSION SIGNATURE OF SCLC BASED ON AUTOPSY SPECIMENS A patient (64-year-old Japanese man) who died of SCLC underwent an autopsy. The patient had an
excessive tobacco-smoking custom (90 pack years). His father had also died of lung cancer. Immunohistochemical examination demonstrated consistent expression of the neuron-specific antigen
(synaptophysin and CD56). Specimens were obtained from primary lung lesions and metastatic liver and brain lesions. The clinical course of the patient is summarized in Figure 1. Expression
of miRNA patterns were analyzed by the TaqMan LDA Human microRNA Panel v2.0 (Applied Biosystems, Foster City, CA, USA). The assay procedure was described previously.14 A cutoff _P_-value of
<0.05 was used to narrow down the candidates after global normalization of the raw data. After global normalization, additional normalization was carried out with _RNU48_. CELL LINES, RNA
ISOLATION Human SCLC cell lines (SBC-3 and NCI-H466 cells) were obtained from the Japanese Cancer Research Resources Bank (Osaka, Japan) and the American Type Culture Collection (Manassas,
VA, USA), respectively. Total RNA was isolated from cultured cells using Isogen (Nippon Gene, Tokyo, Japan). Total RNA was obtained from formalin-fixed, paraffin-embedded human clinical
specimens using Recover All Total Nucleic Acid Isolation kit (Ambion, Austin, TX, USA) as described previously.18, 19, 20 QUANTITATIVE REAL-TIME REVERSE TRANSCRIPTION-PCR The procedure for
PCR quantification was described previously.18, 19, 20 Stem–loop reverse transcription-PCRs for _miR-27a-5p_ (P/N: 002445; Applied Biosystems), _miR-485-3p_ (P/N: 001277), _miR-34b-3p_ (P/N:
002102) and _miR-574-3p_ (P/N: 002349) were used in this study. Human _GUSB_ (P/N: Hs99999908_m1; Applied Biosystems) or _RNU48_ (P/N: 001006; Applied Biosystems) were used to normalize the
data for quantification of mRNA and miRNAs, respectively. TRANSFECTION WITH MIRNA MIMIC AND CELL PROLIFERATION, MIGRATION AND INVASION ASSAYS The following mature miRNA species were used in
the present study: Pre-miR miRNA precursors (_hsa-miR-27a-5p_, P/N: AM 13096; _hsa-miR-485-3p_, P/N: AM 10799; _hsa-miR-34b-3p_, P/N: AM 12727; _hsa-miR-574-3p_, P/N: AM 12848; Applied
Biosystems) and negative control miRNA, P/N: AM 17111; Applied Biosystems. RNAs were incubated with OPTI-MEM (Invitrogen, Carlsbad, CA, USA) and Lipofectamine RNAiMAX reagent (Invitrogen) as
described previously.18, 19, 20 Cells were transfected with 10 nm miRNAs by reverse transfection as described previously.18, 19, 20 Cell migration assays and cell invasion assays were
performed using modified Boyden chambers with 8 μm pores in 24-well tissue culture plates. Chambers for cell invasion assays consisted of Transwell-precoated Matrigel membrane filter inserts
(BD Biosciences, Bedford, MA, USA). After 48 h of transfection, cells were plated in 24-well plates at 4 × 105 cells (SBC-3 cells) and 2 × 105 cells (NCI-H446 cells) per well, respectively.
All experiments were performed in three independent trials. IDENTIFICATION OF ONCOGENIC GENES TARGETED BY TUMOR-SUPPRESSIVE MIRNAS To identify genes putatively targeted by _miR-27a-5p_ and
_miR-34b-3p_, we performed _in silico_ analysis using the TargetScan database and GEO expression data. First, we screened _miR-27a-5p_- and _miR-34b-3p_-targeted genes using the TargetScan
database (release 7.1: http://www.targetscan.org/vert_71/). Next, we paired down the lists of genes based on a publicly available gene expression data set in a GEO database (accession
number: GSE43346). Finally, we identified common genes targeted by _miR-27a-5p_ and _miR-34b-3p_. IMMUNOHISTOCHEMISTRY Three tissue autopsy specimens were used: SCLC lung tissue in the
primary lesion and two metastatic tissues from the liver and brain. Specimens were immunostained following the manufacturer's protocol with the Ultra-Vision Detection System (Thermo
Scientific, Fremont, CA, USA). For immunohistochemistry, we used primary rabbit polyclonal antibodies against the following: TOP2A (1:200, HPA006458; Sigma-Aldrich, St Louis, MO, USA), MELK
(1:200, HPA017214; Sigma-Aldrich), CENPF (1:400, ab5; Abcam, Cambridge, UK) and SOX1 (1:500, ab87775; Abcam). The procedure was carried out as described previously.18, 19 STATISTICS The
relationships between two groups and the numerical values obtained by real-time reverse transcription-PCR were analyzed using Mann–Whitney _U_-tests. The relationships among more than three
variables and numerical values were analyzed using the Bonferroni-adjusted Mann–Whitney _U_-test. RESULTS CONSTRUCTION OF THE MIRNA EXPRESSION SIGNATURE OF SCLC SPECIMENS Using a PCR-based
array system, we analyzed differentially expressed miRNAs in the primary SCLC lesion and metastatic lesions (liver and brain) and compared them with non-cancerous lesions. Based on this
analysis, we listed the top 35 downregulated miRNAs in SCLC tissues (Table 2). Among them, we focused on four miRNAs (_miR-27a-5p_, _miR-485-3p, miR-34b-3p_ and _miR-574-3p_) that were
significantly downregulated in SCLC specimens. EXPRESSION LEVELS OF _MIR-27A-5P_, _MIR-485-3P, MIR-34B-3P_ AND _MIR-574-3P_ IN LUNG CANCER CLINICAL SPECIMENS AND SCLC CELL LINES We evaluated
the expression levels of four downregulated miRNAs (_miR-27a-5p_, _miR-485-3p, miR-34b-3p_ and _miR-574-3p_) in non-cancerous (_n_=27), SCLC (_n_=11) and NSCLC (_n_=52) clinical specimens
and in cell lines (SBC-3 and NCI-H446). The expression levels of _miR-27a-5p_ and _miR-34b-3p_ were significantly reduced in SCLC specimens compared with non-cancerous specimens (Figure 2).
These miRNAs expression levels were also low in both SCLC cell lines (SBC-3 and NCI-H446). EFFECTS OF RESTORING _MIR-27A-5P_, _MIR-485-3P, MIR-34B-3P_ AND _MIR-574-3P_ ON CELL PROLIFERATION,
MIGRATION AND INVASION IN SCLC CELL LINES To investigate the functional roles of miRNAs in SCLC, we performed gain-of-function studies in SBC-3 and NCI-H446 cells by transfecting the cells
with miRNA mimics. XTT assays showed that cell proliferation was significantly inhibited in _miR-27a-3p_ transfectants of SBC-3 in comparison with the mock, although inhibition was not seen
in NCI-H446. Additionally _miR-34b-3p_ transfection significantly inhibited the proliferation of both cell lines in comparison with mock. Furthermore, cell proliferation was not inhibited in
_miR-485-3p_ and _miR-574-3p_ transfectants of SBC-3 compared with mock, whereas it was significantly inhibited in NCI-H446 transfectants (Figure 3a). _In vitro_ assays showed that cell
migration was inhibited in _miR-485-3p_ transfectants of SBC-3 in comparison with the mock, whereas it was not inhibited in NCI-H446 transfectants. _miR-34b-3p_ transfection significantly
inhibited cell migration in both cell lines compared with mock. Cell migration activity was not inhibited in _miR-574-3p_ transfectants of SBC-3 compared with mock, whereas it was
significantly inhibited in NCI-H446 transfectants (Figure 3b). Finally, Matrigel invasion assays demonstrated that cell invasive activity was significantly inhibited in _miR-27a-3p_,
_miR-485-3p_ and _miR-574-3p_ transfectants of SBC-3 in comparison with the mock. Inhibition was not seen in NCI-H446 transfectants. Finally, _miR-34b-3p_ transfection significantly
inhibited cell invasion in both cell lines (Figure 3c). IDENTIFICATION OF PUTATIVE TARGET GENES REGULATED BY _MIR-27A-5P_ AND _MIR-34B-3P_ IN SCLC To identify putative target genes subjected
to _miR-27a-5p_ and _miR-34b-3p_ regulation, we performed _in silico_ analysis using the TargetScan database and GEO expression data. First, we screened _miR-27a-5p_- and
_miR-34b-3p_-targeted genes using the TargetScan database (release 7.1: http://www.targetscan.org/vert_71/). We found 3238 and 4165 genes that had putative target sites in their 3′-UTRs for
_miR-27a-5p_ and for _miR-34b-3p_, respectively. Second, we paired down the lists of genes based on a publicly available gene expression data set in the GEO database (accession number:
GSE43346) and identified 7 and 17 genes, respectively. Finally, we selected the following four common genes from those lists: _TOP2A_, _MELK_, _CENPF_ and _SOX1._ The flow chart outlining
our strategy for identification of putative target genes of _miR-27a-5p_ and _miR-34b-3p_ is shown in Figure 4. IMMUNOHISTOCHEMICAL STAINING OF MIR-TARGETED PROTEINS (TOP2A, MELK, CENPF AND
SOX1) IN SCLC CLINICAL SPECIMENS To validate expression of TOP2A, MELK, CENPF and SOX1 proteins, immunohistochemistry was used to assess SCLC autopsy specimens. Immunohistochemical staining
demonstrated overexpression of TOP2A in the nuclei of the primary lesion and liver metastasis (Figure 5a). The expression of MELK was high in the cytoplasm of all sites (Figure 5b). Also,
the expression of CENPF was relatively high in the nuclei of the primary lesion (Figure 5c). However the expression of SOX1 was not observed in any site. Therefore, it appeared that _TOP2A_,
_MELK_ and _CENPF_ play key roles as oncogenes regulated by _miR-27a-5p_ and _miR-34b-3p. TOP2A_ and _MELK_ appeared to be particularly important in the development of SCLC. DISCUSSION SCLC
is a highly aggressive cancer with a poor prognosis because most patients are diagnosed with extensive disease.1, 2 Treatment strategies for late stage SCLC are commonly platinum-based
chemotherapy.3, 4 Although SCLC cells are initially very sensitive to cytotoxic chemotherapy, in most cases, SCLC cells acquire resistance to these treatments.5 No effective treatments are
approved for recurrence and the distant metastases of the disease. To improve the dismal prognosis of SCLC, effective treatment strategies are urgently needed. Uncovering the genomic
characteristics of the disease might add new therapeutic targets. A growing body of evidence has shown that dysregulated miRNAs are deeply involved in human oncogenesis, metastasis and drug
resistance.11 Aberrantly expressed miRNAs can destroy tightly controlled RNA networks and promote oncogenic development. In this study, we first identified dysregulated miRNAs based on miRNA
expression signatures of autopsy specimens from a patient with SCLC. The patient had undergone several therapeutic treatments, thus we considered that these specimens were likely
treatment-resistant SCLC cells. Our present data showed that several miRNAs, such as _miR-27a-5p_, _miR-485-3p, miR-34b-3p_ and _miR-574-3p,_ were markedly downregulated in cancerous tissues
based on profiles of SCLC autopsy specimens. Among these miRNAs, clinical specimens confirmed that the expression levels of _miR-27a-5p_ and _miR-34b-3p_ were significantly reduced in SCLC
tissues compared with non-cancerous tissues. Additionally, ectopic expression of these two miRNAs significantly inhibited cancer cell aggressiveness, suggesting that _miR-27a-5p_ and
_miR-34b-3p_ functioned as tumor suppressors in SCLC cells. Based on these results, we focused on these two miRNAs and explored the molecular networks that they regulated. By miRNA database
searching (miRBase; http://www.mirbase.org/), pre-_miR-27a_ produces two types of mature miRNAs, _miR-27a-5p_ and _miR-27a-3p._ The _miR-27a-5p_ is passenger strand of pre-_miR-27a_, whereas
_miR-27a-3p_ is guide strand of it. Most of past articles focused on the functional significance of the _miR-27a-3p_ in several cancers. The functional significance of _miR-27a-3p_ has
confusion in cancer cells, including lung cancer. Previous studies have shown that _miR-27a-3p_ is frequently upregulated and plays functional roles in multiple tumor types, including
pancreatic cancer, breast cancer, ovarian cancer, esophageal cancer, renal cell carcinoma, hepatocellular carcinoma, glioma and gastric cancer.21 The guide strand of _miR-27a-3p_ promotes
tumorigenesis in several types of cancer.21, 22 On the other hand, _miR-27a-3p_ was directly regulated tyrosine kinase receptors, EGFR and MET in lung cancer.23 Cancer stem cells is a
promising target for cancer therapy in cases of cancer cell aggressiveness and drug resistance. Downregulation of _miR-27a-3p_ was observed in sphere-forming cells in SCLC.24 Inhibition of
_miR-27a-3p_ in parental cells enhanced stem-like properties of SCLC cells _in vitro_.24 The passenger strand of pre-_miR-27a (miR-27a-5p)_ is downregulated in head and neck squamous cell
carcinoma, and it acts as a tumor suppressor by targeting the EGFR signaling axis. _miR-27a-5p_ simultaneously decreases expression of _EGFR_, _AKT1_ and _mTOR,_ leading to decreased solid
tumor viability.22 In the past established theory of miRNA biogenesis, the passenger strand of miRNA is degradation and not incorporated into RNA-induced silencing complex.7 Surprisingly,
our recent studies showed that _miR-145-3p_ (passenger stand of pre_-miR-145_) actually functioned as anti-tumor miRNA in lung cancer and bladder cancer.25, 26 Similarly, we confirmed the
anti-tumor function of _miR-139-3p_ (passenger strand of pre-_miR-139_) in bladder cancer.27 These findings indicate that some passenger strand of miRNAs have biological function in cells.
The involvement of passenger strand miRNAs in the regulation of cellular processes is a novel concept in RNA research. It is an important study theme to investigate molecular mechanisms of
transcriptional control of miRNAs in cancer cells. Silencing mechanisms of _miR-27a-5p_ remain largely undefined. A recent study showed that Twist-1, a transcription factor of
epithelial–mesenchymal transition regulation, was negatively controlled by _miR-27a-3p_ expression in hepatocellular carcinoma.28 Other study showed that hepatocyte growth factor-mediated
MET signal induced _miR-27a-3p_ expression in lung cancer cells.23 The further study is necessary to elucidate expression control of _miR-27a-5p_ in SCLC. The expression level of
_miR-34b-3p_ is decreased in several cancers, such as neuroblastoma and cervical cancer.29, 30 In neuroblastoma, _miR-34b-3p_ functions as a tumor suppressor by targeting _CCNE2_ and
_E2F3_.29 A representative tumor suppressor p53 regulated the expression control of several miRNAs. The _miR-34_-family (_miR-34a/b/c_) was direct targets of p53 regulation.31 Expression of
the _miR-34_-family caused antitumor effects, such as inducing apoptosis and cell cycle arrest. Therefore, cancer cells enhance inactivation of _miR-34_-family expression through CpG
methylation.31 However, the functions of these miRNAs are still not fully understood. Based on those studies and our findings, we investigated the molecular networks regulated by the miRNAs
that we identified to better understand the etiology of SCLC. We hypothesized that _miR-27a-5p_ and _miR-34b-3p_ might coordinately regulate target genes associated with SCLC pathogenesis.
Therefore, we performed _in silico_ analysis and identified four genes (_TOP2A_, _MELK_, _CENPF_ and _SOX1_) that were potential targets of _miR-27a-5p_ and _miR-34b-3p_.
Immunohistochemistry indicated that MELK, TOP2A and CENPF play key roles in promoting oncogenesis. We suggest that TOP2A and MELK are particularly important in SCLC. MELK is classified as a
member of the SLK/AMPK serine–threonine kinase family and known as an embryonic and neural stem cell marker. It is associated with cell survival, proliferation and apoptosis in various
cancers.32, 33 Several studies have reported a correlation between _MELK_ gene expression and tumor malignancy grade for astrocytoma and breast cancer.34, 35 It was reported that _MELK_
could play roles in cell cycle regulation (possibly in G0–G1 and S phases) as well as in responses against radiation and 5-FU treatment in colorectal cancer cells.32 Inoue _et al._36
demonstrated that _MELK_ was highly expressed in most SCLC cell lines and primary SCLC tumors. In this study, we demonstrated that the cancerous tissues of autopsy specimens that were
presumably resistant to drug therapies stained strongly for MELK, suggesting that _MELK_ was involved in resistance to chemotherapy in SCLC. TOP2A is a subfamily of DNA topoisomerase type II
that controls and alters the topologic states of DNA during transcription. Abnormal alterations of _TOP2A_ include changes in gene copy number and gene expression level in cancer cells.37
Aberrant expression of _TOP2A_ is generally associated with poor prognosis in breast cancer, ovarian cancer, oral cancer, esophageal cancer and lung cancer.38, 39, 40 In particular,
multidrug resistance protein and TOP2A are involved in drug resistance in NSCLC.41 We confirmed the upregulation of _TOP2A_ in SCLC autopsy specimens by immunohistochemistry. CENPF encodes a
protein that associates with the centromere–kinetochore complex. This protein is a member of the centromere protein family and acts in a critical chromosomal segregation process, including
kinetochore assembly and spindle checkpoint signaling during mitosis.42 Overexpression of _CENPF_ has been observed in prostate cancer and breast cancer.42, 43 Our recent study showed that
downregulated tumor-suppressive _miR-205_ enhanced prostate cancer aggressiveness through direct regulation of _CENPF_.42 In conclusion, the expression of _miR-27a-5p_ and _miR-34b-3p_ was
downregulated in SCLC autopsy specimens. They both act as tumor suppressors in SCLC cells. Oncogenic _MELK_, _TOP2A_ and _CENPF_ are regulated by these miRNAs, and high expression of those
oncogenes in SCLC autopsy specimens indicates clinical importance. Elucidation of _miR-27a-5p-_ and _miR-34b-3p_-mediated molecular networks may lead to a better understanding of SCLC
aggressiveness and the development of new treatment strategies. ACCESSION CODES ACCESSIONS GENE EXPRESSION OMNIBUS * GSE43346 REFERENCES * Chute, J. P., Chen, T., Feigal, E., Simon, R. &
Johnson, B. E. Twenty years of phase III trials for patients with extensive-stage small-cell lung cancer: perceptible progress. _J. Clin. Oncol._ 17, 1794–1801 (1999). Article CAS Google
Scholar * Lassen, U., Osterlind, K., Hansen, M., Dombernowsky, P., Bergman, B. & Hansen, H. H. Long-term survival in small-cell lung cancer: posttreatment characteristics in patients
surviving 5 to 18+ years—an analysis of 1714 consecutive patients. _J. Clin. Oncol._ 13, 1215–1220 (1995). Article CAS Google Scholar * Amarasena, I. U., Walters, J. A., Wood-Baker, R.
& Fong, K. Platinum versus non-platinum chemotherapy regimens for small cell lung cancer. _Cochrane Database Syst. Rev._ 8, CD006849 (2015). Google Scholar * Satouchi, M., Kotani, Y.,
Shibata, T., Ando, M., Nakagawa, K., Yamamoto, N. _et al_. Phase III study comparing amrubicin plus cisplatin with irinotecan plus cisplatin in the treatment of extensive-disease small-cell
lung cancer: JCOG 0509. _J. Clin. Oncol._ 32, 1262–1268 (2014). Article CAS Google Scholar * van Meerbeeck, J. P., Fennell, D. A. & De Ruysscher, D. K. Small-cell lung cancer.
_Lancet_ 378, 1741–1755 (2011). Article Google Scholar * Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. _Cell_ 136, 642–655 (2009). Article CAS
Google Scholar * Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. _Cell_ 116, 281–297 (2004). Article CAS Google Scholar * Filipowicz, W., Bhattacharyya, S. N.
& Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? _Nat. Rev. Genet._ 9, 102–114 (2008). Article CAS Google Scholar * Friedman, R.
C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. _Genome Res._ 19, 92–105 (2009). Article CAS Google Scholar * Nelson, K. M.
& Weiss, G. J. MicroRNAs and cancer: past, present, and potential future. _Mol Cancer Ther_ 7, 3655–3660 (2008). Article CAS Google Scholar * Wiemer, E. A. The role of microRNAs in
cancer: no small matter. _Eur. J. Cancer._ 43, 1529–1544 (2007). Article CAS Google Scholar * Itesako, T., Seki, N., Yoshino, H., Chiyomaru, T., Yamasaki, T., Hidaka, H. _et al_. The
microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. _PLoS ONE_ 9, e84311 (2014). Article Google Scholar * Fukumoto,
I., Kinoshita, T., Hanazawa, T., Kikkawa, N., Chiyomaru, T., Enokida, H. _et al_. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. _Br. J. Cancer_ 111, 386–394 (2014). Article CAS Google Scholar * Moriya, Y., Nohata, N., Kinoshita, T., Mutallip, M., Okamoto, T., Yoshida, S. _et al_.
Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. _J. Hum. Genet._ 57, 38–45 (2012). Article CAS Google Scholar * Goto, Y., Kojima, S.,
Nishikawa, R., Kurozumi, A., Kato, M., Enokida, H. _et al_. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour
suppressor and disease progression marker. _Br. J. Cancer_ 113, 1055–1065 (2015). Article CAS Google Scholar * Goto, Y., Kurozumi, A., Nohata, N., Kojima, S., Matsushita, R., Yoshino, H.
_et al_. The microRNA signature of patients with sunitinib failure: regulation of UHRF1 pathways by microRNA-101 in renal cell carcinoma. _Oncotarget_ 7, 59070–59096 (2016). PubMed PubMed
Central Google Scholar * Shepherd, F. A., Crowley, J., Van Houtte, P., Postmus, P. E., Carney, D., Chansky, K. _et al_. The International Association for the Study of Lung Cancer Lung
Cancer Staging Project: proposals regarding the clinical staging of small cell lung cancer in the forthcoming (seventh) edition of the tumor, node, metastasis classification for lung cancer.
_J. Thorac. Oncol._ 2, 1067–1077 (2007). Article Google Scholar * Mataki, H., Seki, N., Chiyomaru, T., Enokida, H., Goto, Y., Kumamoto, T. _et al_. Tumor-suppressive microRNA-206 as a
dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. _Int. J. Oncol._ 46, 1039–1050 (2015). Article CAS Google Scholar * Kamikawaji, K., Seki, N., Watanabe,
M., Mataki, H., Kumamoto, T., Takagi, K. _et al_. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. _J. Hum. Genet._ 61, 985–993
(2016). Article CAS Google Scholar * Mataki, H., Enokida, H., Chiyomaru, T., Mizuno, K., Matsushita, R., Goto, Y. _et al_. Downregulation of the microRNA-1/133a cluster enhances cancer
cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. _J. Hum. Genet._ 60, 53–61 (2015). Article CAS Google Scholar * Zhou, L., Liang, X., Zhang, L.,
Yang, L., Nagao, N., Wu, H. _et al_. MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. _Oncotarget_ 7, 51943–51954 (2016). PubMed PubMed Central Google Scholar *
Wu, X., Bhayani, M. K., Dodge, C. T., Nicoloso, M. S., Chen, Y., Yan, X. _et al_. Coordinated targeting of the EGFR signaling axis by microRNA-27a*. _Oncotarget_ 4, 1388–1398 (2013). PubMed
PubMed Central Google Scholar * Acunzo, M., Romano, G., Palmieri, D., Lagana, A., Garofalo, M., Balatti, V. _et al_. Cross-talk between MET and EGFR in non-small cell lung cancer
involves miR-27a and Sprouty2. _Proc. Natl Acad. Sci. USA_ 110, 8573–8578 (2013). Article CAS Google Scholar * Miao, Y., Li, J., Qiu, X., Li, Y., Wang, Z. & Luan, Y. miR-27a regulates
the self renewal of the H446 small cell lung cancer cell line _in vitro_. _Oncol. Rep._ 29, 161–168 (2013). Article CAS Google Scholar * Mataki, H., Seki, N., Mizuno, K., Nohata, N.,
Kamikawaji, K., Kumamoto, T. _et al_. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. _Oncotarget_ 7,
72084–72098 (2016). Article Google Scholar * Matsushita, R., Yoshino, H., Enokida, H., Goto, Y., Miyamoto, K., Yonemori, M. _et al_. Regulation of UHRF1 by dual-strand tumor-suppressor
microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. _Oncotarget_ 7, 28460–28487 (2016). Article Google Scholar * Yonemori, M., Seki, N., Yoshino,
H., Matsushita, R., Miyamoto, K., Nakagawa, M. _et al_. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. _Cancer Sci._ 107, 1233–1242
(2016). Article CAS Google Scholar * Zhao, N., Sun, H., Sun, B., Zhu, D., Zhao, X., Wang, Y. _et al_. miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin
expression and inhibiting EMT: an essential role for Twist-1 in HCC. _Sci. Rep._ 6, 23091 (2016). Article CAS Google Scholar * Maugeri, M., Barbagallo, D., Barbagallo, C., Banelli, B., Di
Mauro, S., Purrello, F. _et al_. Altered expression of miRNAs and methylation of their promoters are correlated in neuroblastoma. _Oncotarget_ 7, 83330–83341 (2016). Article Google Scholar
* Revathidevi, S., Manikandan, M., Rao, A. K., Vinothkumar, V., Arunkumar, G., Rajkumar, K. S. _et al_. Analysis of APOBEC3A/3B germline deletion polymorphism in breast, cervical and oral
cancers from South India and its impact on miRNA regulation. _Tumour Biol._ 37, 11983–11990 (2016). Article CAS Google Scholar * Hermeking, H. The miR-34 family in cancer and apoptosis.
_Cell Death Differ._ 17, 193–199 (2010). Article CAS Google Scholar * Choi, S. & Ku, J. L. Resistance of colorectal cancer cells to radiation and 5-FU is associated with MELK
expression. _Biochem. Biophys. Res. Commun._ 412, 207–213 (2011). Article CAS Google Scholar * Lin, M. L., Park, J. H., Nishidate, T., Nakamura, Y. & Katagiri, T. Involvement of
maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. _Breast Cancer Res._ 9, R17 (2007).
Article Google Scholar * Ganguly, R., Mohyeldin, A., Thiel, J., Kornblum, H. I., Beullens, M. & Nakano, I. MELK-a conserved kinase: functions, signaling, cancer, and controversy.
_Clin. Transl. Med._ 4, 11 (2015). Article Google Scholar * Ganguly, R., Hong, C. S., Smith, L. G., Kornblum, H. I. & Nakano, I. Maternal embryonic leucine zipper kinase: key kinase
for stem cell phenotype in glioma and other cancers. _Mol. Cancer Ther._ 13, 1393–1398 (2014). Article CAS Google Scholar * Inoue, H., Kato, T., Olugbile, S., Tamura, K., Chung, S.,
Miyamoto, T. _et al_. Effective growth-suppressive activity of maternal embryonic leucine-zipper kinase (MELK) inhibitor against small cell lung cancer. _Oncotarget_ 7, 13621–13633 (2016).
PubMed PubMed Central Google Scholar * Chen, T., Sun, Y., Ji, P., Kopetz, S. & Zhang, W. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. _Oncogene_
34, 4019–4031 (2015). Article CAS Google Scholar * Ceppi, P., Longo, M., Volante, M., Novello, S., Cappia, S., Bacillo, E. _et al_. Excision repair cross complementing-1 and topoisomerase
IIalpha gene expression in small-cell lung cancer patients treated with platinum and etoposide: a retrospective study. _J. Thorac. Oncol._ 3, 583–589 (2008). Article Google Scholar *
Meng, H., Chen, R., Li, W., Xu, L. & Xu, L. Correlations of TOP2A gene aberrations and expression of topoisomerase IIalpha protein and TOP2A mRNA expression in primary breast cancer: a
retrospective study of 86 cases using fluorescence _in situ_ hybridization and immunohistochemistry. _Pathol. Int._ 62, 391–399 (2012). Article CAS Google Scholar * Dingemans, A. M.,
Witlox, M. A., Stallaert, R. A., van der Valk, P., Postmus, P. E. & Giaccone, G. Expression of DNA topoisomerase IIalpha and topoisomerase IIbeta genes predicts survival and response to
chemotherapy in patients with small cell lung cancer. _Clin. Cancer Res._ 5, 2048–2058 (1999). CAS PubMed Google Scholar * Huang, H., Liu, J., Meng, Q. & Niu, G. Multidrug resistance
protein and topoisomerase 2 alpha expression in non-small cell lung cancer are related with brain metastasis postoperatively. _Int. J. Clin. Exp. Pathol._ 8, 11537–11542 (2015). CAS PubMed
PubMed Central Google Scholar * Nishikawa, R., Goto, Y., Kurozumi, A., Matsushita, R., Enokida, H., Kojima, S. _et al_. MicroRNA-205 inhibits cancer cell migration and invasion via
modulation of centromere protein F regulating pathways in prostate cancer. _Int. J. Urol._ 22, 867–877 (2015). Article CAS Google Scholar * O'Brien, S. L., Fagan, A., Fox, E. J.,
Millikan, R. C., Culhane, A. C., Brennan, D. J. _et al_. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer. _Int. J.
Cancer_ 120, 1434–1443 (2007). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS The present study was supported by KAKENHI(C) grant 15K10801 and 16K19458. AUTHOR
INFORMATION Author notes * Keiko Mizuno and Hiroko Mataki: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Pulmonary Medicine, Graduate School of
Medical and Dental Sciences, Kagoshima University, Kagoshima, Japan Keiko Mizuno, Hiroko Mataki, Kazuto Kamikawaji, Tomohiro Kumamoto & Hiromasa Inoue * Department of Functional
Genomics, Chiba University Graduate School of Medicine, Chiba, Japan Takayuki Arai, Atsushi Okato & Naohiko Seki * Department of Pathology, Field of Oncology, Graduate School of Medical
and Dental Sciences, Kagoshima University, Kagoshima, Japan Tsubasa Hiraki & Kazuhito Hatanaka Authors * Keiko Mizuno View author publications You can also search for this author
inPubMed Google Scholar * Hiroko Mataki View author publications You can also search for this author inPubMed Google Scholar * Takayuki Arai View author publications You can also search for
this author inPubMed Google Scholar * Atsushi Okato View author publications You can also search for this author inPubMed Google Scholar * Kazuto Kamikawaji View author publications You can
also search for this author inPubMed Google Scholar * Tomohiro Kumamoto View author publications You can also search for this author inPubMed Google Scholar * Tsubasa Hiraki View author
publications You can also search for this author inPubMed Google Scholar * Kazuhito Hatanaka View author publications You can also search for this author inPubMed Google Scholar * Hiromasa
Inoue View author publications You can also search for this author inPubMed Google Scholar * Naohiko Seki View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to Naohiko Seki. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information
accompanies the paper on Journal of Human Genetics website SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1 (XLSX 14 KB) SUPPLEMENTARY TABLE 2 (XLSX 10 KB) RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Mizuno, K., Mataki, H., Arai, T. _et al._ The microRNA expression signature of small cell lung cancer: tumor suppressors of _miR-27a-5p_ and
_miR-34b-3p_ and their targeted oncogenes. _J Hum Genet_ 62, 671–678 (2017). https://doi.org/10.1038/jhg.2017.27 Download citation * Received: 12 January 2017 * Revised: 02 February 2017 *
Accepted: 07 February 2017 * Published: 09 March 2017 * Issue Date: July 2017 * DOI: https://doi.org/10.1038/jhg.2017.27 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative