- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Biochar and mineral-enriched biochar (MEB) have been used as soil amendments to improve soil fertility, sequester carbon and mitigate greenhouse gas emissions. Such beneficial
outcomes could be partially mediated by soil bacteria, however little is known about how they directly interact with biochar or MEB. We therefore analyzed the diversity and functions of
bacterial communities on the surfaces of one biochar and two different MEBs after a 140-day incubation in soil. The results show that the biochar and the MEBs harbor distinct bacterial
communities to the bulk soil. Communities on biochar and MEBs were dominated by a novel Gammaproteobacterium. Genome reconstruction combined with electron microscopy and high-resolution
elemental analysis revealed that the bacterium generates energy from the oxidation of iron that is present on the surface. Two other bacteria belonging to the genus _Thiobacillus_ and a
novel group within the _Oxalbacteraceae_ were enriched only on the MEBs and they had the genetic capacity for thiosulfate oxidation. All three surface-enriched bacteria also had the capacity
to fix carbon dioxide, either in a potentially strictly autotrophic or mixotrophic manner. Our results show the dominance of chemolithotrophic processes on the surface of biochar and MEB
that can contribute to carbon sequestration in soil. SIMILAR CONTENT BEING VIEWED BY OTHERS POTENTIAL OF BIOCHAR TO RESTORATION OF MICROBIAL BIOMASS AND ENZYMATIC ACTIVITY IN A HIGHLY
DEGRADED SEMIARID SOIL Article Open access 30 October 2024 EFFECTS OF MIXED BIOCRUSTS ON SOIL NUTRIENTS AND BACTERIAL COMMUNITY STRUCTURE: A CASE STUDY FROM HILLY LOESS PLATEAU, CHINA
Article Open access 11 September 2024 THE MICROBIAL COMMUNITY FROM THE EARLY-PLANT COLONIZER (_BACCHARIS LINEARIS_) IS REQUIRED FOR PLANT ESTABLISHMENT ON COPPER MINE TAILINGS Article Open
access 17 May 2021 INTRODUCTION Biochar is a carbon-rich solid material derived from the thermal processing of biomass in an oxygen-depleted environment (Lehmann and Joseph, 2015). The
application of biochar to soil has shown promising results for the sequestration of carbon (Lehmann et al., 2006), the mitigation of greenhouse gas emissions (Woolf et al., 2010), the
immobilization of heavy metals (Cao et al., 2011) and the improvement of soil fertility (Kolton et al., 2011). However, biochars often have to be applied in high rates (10–100 t ha−1) to
agricultural soils to deliver such positive outcomes (Jeffery et al., 2011). Recent findings have shown that biochar naturally forms aggregates with minerals in the highly fertile Amazonian
Dark Earths (Chia et al., 2012). This has led to the manufacturing of biochars with coatings of ground rocks, clays and other minerals to form so called mineral-enriched biochar (MEB; Chia
et al., 2014). At low application rates (~5 t ha−1) these MEBs have been found to produce sweet corn yields comparable to traditional fertilizer (Nielsen et al., 2014) and improve
productivity of pakchoi in organic farming (Ye et al., 2016). The beneficial effects of biochar or MEB have been attributed to their recalcitrance, conductivity, porosity and adsorption
properties, which depend on the biomass feedstock, pyrolysis conditions and mineral additives, if used (Chia et al., 2015). However, biochar and MEB have also been shown to cause shifts in
microbial communities (O’Neill et al., 2009; Anderson et al., 2011; Nielsen et al., 2014; Abujabhah et al., 2016) and this might indirectly impact biogeochemical processes in soil (Lehmann
et al., 2011; Bardgett and van der Putten, 2014). For example, a corn stalk biochar has been shown to increase the abundances of methanotrophic proteobacteria in a Chinese paddy soil (Feng
et al., 2012), a jarrah wood MEB increased soil nitrifiers (Ye et al., 2016) and a green waste biochar stimulated N2O-reducing bacteria (Harter et al., 2014). These and other studies have
mainly focused on bulk measurements of amended soils to explain the microbial processes behind the beneficial effects of biochar and MEB, while very few studies have examined the specific
interactions between the biochar surfaces and microorganisms. It has been well recognized that soil microorganisms colonize surfaces of soil particles, which provide specific habitats in
terms of inorganic and organic substrates, oxygen level or redox conditions (Sessitsch et al., 2001; Mills, 2003). Different soil particle fractions often harbor different microbial
communities and hence support distinct metabolic processes (Hemkemeyer et al., 2015). Likewise, biochar has been suggested to provide habitats for microorganisms when added to soils
(Pietikäinen et al., 2000; Lehmann et al., 2011; Quilliam et al., 2013). Indeed, Sun et al. (2016) recently reported for the first time that bacterial communities on biochar particles and
bulk soil do significantly differ. In addition, the study used 16S rRNA gene sequence analysis and a Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
(PICRUSt; Langille et al., 2013) to predict that the bacterial communities on biochar particles prefer to metabolize xenobiotics. However, given that soil microorganisms are highly diverse
and largely uncharacterized (Rondon et al., 1999; Fierer and Jackson, 2006), it is likely that certain metabolic traits of bacterial communities on biochar surface have been missed by such
purely 16S rRNA-based functional predictions. To improve our understanding of the processes that occur on biochar surfaces, we investigate here bacterial communities and their functions on a
bamboo biochar and two biochars enriched with minerals. We hypothesized that specific bacterial communities would be recruited by the different biochars and that they carry out distinct
functions that are determined by specific aspects of the biochar surface. Using amplicon sequencing of the bacterial 16S rRNA gene and metagenomics, we show a substantial surface enrichment
of specific soil bacteria that have the capacity for different kinds of chemolithotrophy based on genome-based predictions. We then use a novel gold-label _in situ_ hybridization (GISH)
method (Ye et al., 2015) and scanning electron microscopy to localize a dominant bacterium on the biochar particles. Using scanning transmission electron microscopy (STEM), energy-dispersive
X-ray spectroscopy (EDS) and electron energy loss spectroscopy (EELS) we show that this dominant bacterium is involved in iron oxidation, which could support CO2 fixation on the biochar
surface. MATERIALS AND METHODS STUDY SOIL, BIOCHAR PRODUCTION AND ANALYSIS Red chromosol soils (Australia Soil Classification) used in this study came from a farmland site located in Dubbo,
central NSW, Australia (32°13′S, 148°59′E). The farmland is within a temperate climate zone with a mean annual rainfall of 584.4 mm and a mean annual temperature of 17.3 °C. The plough layer
(0–20 cm in depth) was collected after crop harvest, homogenized and sieved through a 2 mm mesh. Three different biochars were made from a single length of bamboo to minimize natural
variation in organic chemistry and mineral content. Two of these biochars were treated with minerals and clays to produce MEBs. For this, bamboo was cut into cubes (dimension of 1 cm), which
were randomly divided into three equal parts. Two types of mineral slurries were prepared by dissolving either refined bentonite clay or kaolinite clay (Keane Ceramics, Somersby, Australia)
with ferrous sulfate heptahydrate (FeSO4·7H2O, Sigma-Aldrich, Castle Hill, NSW, Australia) in water at a ratio of 1:1:20 (w/w/w). Bentonite is a smectite with a different crystal structure
and chemical properties to kaolinite, especially in relation to reaction with organic matter (Yariv and Cross, 2001). Our recent study has demonstrated that addition of bentonite produces a
biochar with significantly different carbon structure to that with kaolinite (Rawal et al., 2016). Biochars produced from biomass that has been enriched with iron salts and clays are also
generally much more redox active, store significant quantities of charge under oxygen–starved conditions and have different magnetic properties compared to biochars made from virgin biomass
(Joseph et al., 2015). Two batches of the bamboo cubes were treated by soaking in either of the two slurries at 80 °C for 3 h. These two slurry-treated batches and the untreated batch were
dried at 110 °C for 24 h. Bamboo cubes were then placed into a lab-scale pyrolysis reactor and heated in an oxygen-free environment with a heating rate of 3 °C min−1 to reach 450 °C and kept
there for 30 min. The material was then cooled down and stored under sterile condition until use. These three products were referred to as bamboo biochar (Bam), bentonite biochar (Ben) and
kaolinite biochar (Kao). The chemical compositions of each type of biochar were determined using a vario EL III elemental analyzer (Elementar, Langenselbold, Germany). The dissolved organic
carbon and total soluble nitrogen were extracted with 25 ml of 0.5 m potassium sulfate in an orbital shaker for 1 h at 250 r.p.m. The extracts were then analyzed using a TOC/TNb analyzer
(Analytik Jena, Überlingen, Germany). Results are summarized in Table 1. EXPERIMENTAL DESIGN AND SET-UP Just before application to the soil, the biochar and MEBs were milled into small
pieces (1–5 mm) using sterile mortars and pestles. The biochar and MEBs were applied to soil at a rate of 0.5% w/w (equivalent to 6.5 t ha−1 to a 10 cm soil profile). Mono-ammonium phosphate
was either not added or at a rate of 0.1% w/w (equivalent to 130 kg ha−1 N and 284.7 kg ha−1 P to a 10 cm soil profile) to represent ‘real-world’ agricultural applications of biochar, which
can occur with or without additional fertilization (Nielsen et al., 2014). This crossed design, including controls, resulted in eight treatments: no biochar, Bam, Ben or Kao, each with or
without mono-ammonium phosphate. Two hundred grams of soil were mixed with the corresponding amounts of milled biochar/MEBs and/or fertilizer on a sterile working bench before being placed
into pots (top side 5 cm, bottom side 4 cm and height 12 cm). Triplicate pots for each of the eight treatments were randomly arranged in a 6 × 4 array on a bench within the greenhouse
facility of the University of New South Wales (UNSW). The water content was maintained at 50% water holding capacity during the incubation. The incubation was stopped after 140 days. This
incubation period was chosen to reflect a reasonable approximate of a crop cycle (4–5 months) and because the mean residence time of the labile carbon pool of biochar was found to be about
108 days (Wang et al., 2016). This latter fact means that microbial processes on the biochar and MEB will likely not anymore be influenced by variable leaching of carbon from the biochar. To
process samples after incubation, the content of each pot was poured into individual sterile petri dishes. Biochar particles were manually picked out with sterile forceps. An aliquot of
soil without biochar particles was also taken from this. Biochar particles were transferred onto a sterile 1 mm metal mesh and washed with sterile deionized water to remove attached soil.
Soil and biochar particles were stored at −80 °C until further use. BACTERIAL 16S RRNA GENE ANALYSIS Total DNA was extracted from soil and biochar particles using the PowerSoil DNA isolation
kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instruction, but with minor modification for biochar particles. The kit has a loading capacity of 0.25 g for
soil, but the biochar particles have a low density and hence an equivalent weight could not fit into the tube. Therefore 0.1 g of biochar particles were used, which gave sufficient DNA
yields for subsequent analysis. Extracted DNA was checked for quality and quantity using agarose gel electrophoresis and a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). The V1–V3
regions of the 16S rRNA gene were amplified using the primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 519R (5′-GWATTACCGCGGCKGCTG-3′) that target conserved sequences found in bacteria.
Amplicons from each PCR sample were normalized to equimolar amounts and sequenced using 2 × 300 bp chemistry on a MiSeq platform (Illumina, San Diego, CA, USA) at the Ramaciotti Centre for
Genomics (UNSW). 16S rRNA sequencing data was processed using the MOTHUR MiSeq pipeline (Kozich et al., 2013) and details are provided in the Supplementary Information. In total 2 180 232
high-quality 16S rRNA sequences were generated for 36 samples. After subsampling each sample to an equal sequencing depth (60 562 reads per sample) and clustering, 14 244 operational
taxonomic units (OTUs) at 97% identity were obtained, with the number of OTUs ranging from 1193 to 3988 per sample. The Good’s coverage for the observed OTUs was 99.46±0.02% (mean±s.e.m.)
and the rarefaction curves showed clear asymptotes (Supplementary Figure 1), which together indicate a near-complete sampling of the community. STATISTICAL ANALYSIS The experimental design
consisted of three factors, including biochar type (no biochar, Bam, Ben and Kao), fertilizer (F, NF) and sample type (soil, particle). The bacterial β-diversity was compared using the
Bray–Curtis similarity coefficient calculated on square-root transformed, relative abundances of OTUs and the resulting dissimilarity matrix was mapped using non-metric multidimensional
scaling in the vegan package of R (Oksanen et al., 2015). Permutational analysis of variance (PERMANOVA, using 104 permutations) was applied to the Bray–Curtis dissimilarity matrix to test
the significance levels of differences for each experimental factor using PRIMER V6 (Anderson et al., 2008). The Welch’s t-test within STAMP (Parks et al., 2014) was used to identify OTUs
that showed significant differences in abundance between groups (confidence interval method). Unless otherwise indicated, _P_-values were adjusted for multiple comparisons using the Storey
false discovery rate and displayed as _q_-value (Krzywinski and Altman, 2014). METAGENOMIC SEQUENCING AND ASSEMBLY After checking for quality and quantity of community DNA extracted from
biochar particles, three samples from the treatments of Ben–F–particle, Kao–NF–particle and Kao–F–particle had sufficient high-quality DNA suitable for metagenomic sequencing. Sequencing
libraries for these samples were generated with the NexteraXT kit following the manufacturer’s instructions (Illumina). The libraries were sequenced with an Illumina NextSeq 500 sequencer
using 150-bp paired-end reads at the Australian Centre for Ecogenomics (The University of Queensland, Australia). Approximately 66.5 million read pairs were generated on average per sample
after filtering by PRINSEQ (Schmieder and Edwards, 2011). The metagenomic reads for each sample were assembled into contigs using IDBA-UD with iterative _k_-values setting from 80 to 100
(Peng et al., 2012). All contigs were submitted to the Integrated Microbial Genomes/Expert Review (IMG/ER) system for gene calling and annotation (Markowitz et al., 2012). BINNING AND
ANNOTATION OF DRAFT GENOMES Sequencing reads of all three samples were mapped to contigs longer than 1 kb using Bowtie2 (Langmead and Salzberg, 2012). These contigs were then grouped into
genome bins on the basis of coverage and tetranucleotide frequency using MetaBat (Kang et al., 2015) with the ‘very specific’ option to minimize contaminations. Completeness and
contamination of genome bins were assessed using CheckM (Parks et al., 2015). The genome completeness was estimated by calculating the number of conserved single-copy marker genes recovered
from individual genomic bins. Genome contamination was evaluated from the number of multi-copy marker genes. Genome bins that were greater than 80% completed and with less than 4%
contamination were considered in this study, which resulted in 11 genome bins. Phylosift were used to evaluate taxonomy of each genome bin (Darling et al., 2014). All genome bins were
submitted to the IMG/ER for annotation and gene calling. PHYLOGENETIC ANALYSIS FOR OTU0001 AND OTU0017 The representative sequences of the Gammproteobacterium OTU0001 and _Oxalobacteraceae_
OTU0017 were extracted from the amplicon sequence data. Separately for each OTU, we retrieved 16S rRNA gene sequences (>1300 nt) of the top 50 most closely related type strains from the
nucleotide (nt) database at the National Center for Biotechnology Information (NCBI) using BLASTN (Morgulis et al., 2008) and aligned those with the corresponding OTU sequence using the SINA
web aligner (Pruesse et al., 2012). A phylogenetic tree was constructed using the neighbor joining algorithm in the ARB software package with Jukes-Cantor distance correction (Westram et
al., 2011) and its robustness was tested with 1000 bootstraps. We also retrieved 16S rRNA gene sequences of uncultured organisms that very closely related to OTU0001/OTU0017 via BLASTN from
the nt database. Only sequences with explicit descriptions of their sources were retained and these were inserted into the phylogenetic tree using the parsimony insertion method (Westram et
al., 2011). CLASSIFICATION OF RUBISCO PROTEINS The genomes of the three bacteria dominating the surface community of biochar contained genes encoding for the ribulose-1,5-bisphosphate
carboxylase/oxygenase (RuBisCO) and these were further investigated here. Functionally validated reference sequences were obtained from the scientific literature to determine the phylogeny
of the RuBisCO large subunit genes (_rbcL_). Form II, III and IV RbcL sequences were obtained from Tabita et al. (2008), while form IA, IBc and IC RbcL sequences were from Badger and Bek
(2008). The extracted RbcL sequences from the genome bins were aligned against those reference sequences using ClustalX (Larkin et al., 2007), and then manually curated using Mega (Kumar et
al., 2008). A maximum likelihood phylogenetic tree was constructed using FastTree with a generalized time-reversible model (Price et al., 2010). The confidence level of the tree topology was
evaluated by bootstrap analysis using 1000 sequence replications. NUCLEOTIDE ACCESSION NUMBERS All raw sequencing data sets of this study have been deposited in NCBI Sequence Read Archive.
Amplicon sequences of the 16S rRNA genes were deposited in NCBI BioProject PRJNA313136. The shotgun sequences are available through accession numbers SRR3569623, SRR3569832 and SRR3569833
under PRJNA313136. The annotations of assembled contigs are accessible under the IMG Genome ID 3300005260, 3300005258, 3300005238. The genomic bins of Gama1 (_Gammaproteobacterium_), Oxal1
(_Oxalobacteraceae_) and Thio1 (_Thiobacillus_) are accessible under the IMG Genome ID 2627853547, 2627853544 and 2627853545, respectively. LOCALIZATION OF OTU0001 ON BIOCHAR PARTICLES
OTU0001 was detected using GISH. Briefly, biochar particles were washed in phosphate buffered saline (PBS, 20 mm NaH2PO4, 150 mm NaCl, 1 mm EDTA, pH 6.5) and fixed in 4% (v/v)
paraformaldehyde at 4 °C for 24 h. Fixed biochar particles were then washed in PBS and stored in 1:1 mixture of PBS and absolute ethanol at −20 °C. A nanogold-labeled probe specific for
OTU0001 (see Supplementary Information) was hybridized with the fixed biochar particles. Biochar particles were dehydrated and coated with an evaporated carbon layer (JEE-420 Evaporative
Carbon Coater, JEOL, Peabody, MA,USA). Images of secondary electrons and backscattered electrons were generated with a JEOL 7001F field emission scanning electron microscopy (JEOL, Freising,
Germany). Elemental analyses were conducted using a JEOL silicon drift EDS. Details are provided in the Supplementary Information. COMPOSITIONAL AND OXIDATION STATE ANALYSIS OF PARTICLES IN
CONTACT WITH OTU0001 A GISH-treated biochar particle was first lightly ground into small pieces in absolute ethanol and then transferred onto a lacey carbon support film on a copper grid
inside a JEM-ARM200F aberration-corrected STEM (JEOL, Japan). After the specimen was air dried, initial imaging in bright-field and high-angle annular dark-field (HAADF) were acquired at 200
kV accelerating voltage in a scanning mode using a 5C (0.155 nA) probe and a 40 μm condenser aperture. A NORAN system 7 X-ray Microanalysis System (Thermo Fisher, Fremont, CA, USA) coupled
with a large area (1 steradian) JEOL EDS detector was used for compositional spectral imaging (SI) mapping and a GIF Quantum Energy Filter (Gatan, Warrendale, PA, USA) was used for oxidation
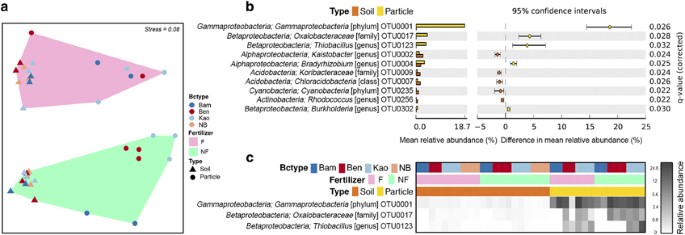
state analyses based on an EELS. RESULTS AND DISCUSSIONS ENRICHMENT OF BACTERIA ON THE SURFACE OF BIOCHAR The factors of biochar type, fertilizer and sample type significantly influenced
bacterial β-diversity as shown in an non-metric multidimensional scaling plot (Figure 1a). PERMANOVA detected significant effects associated with the interactions between biochar type and
fertilizer (_P_=0.03), as well as fertilizer and sample type (_P_=0.04). There were neither significant interactions between biochar type and sample type (_P_=0.34), nor among these three
factors (_P_=0.66; Supplementary Table 1). Together these results show that (i) biochar at a 6.5 t ha−1 application rate, with or without fertilizers, had no impact on the bacterial
community composition of bulk soil over a 140 days incubation, (ii) the addition of fertilizer had a significant impact on bacterial communities in soil and the biochar particle, (iii) the
bacterial communities on the biochar particles were significantly different from those in bulk soils irrespective if fertilizer was present or not and (iv) the bacterial communities on the
two types of MEB particles were significantly different from Bam particles when no fertilizers were applied, however this distinction was disrupted when fertilizer was present. We then
investigated the bacterial taxa that accounted for the observed differences between biochar particles and bulk soils (Figure 1a). Independent of the variation caused by fertilizer
application and biochar type, there were several bacterial taxa that accounted for these differences. In particular three OTUs were dominating the communities found on biochar particles and
were in very low abundance in bulk soils (Figure 1b). These three OTUs could be taxonomically assigned to the class Gammaproteobacteria (OTU0001), the family _Oxalobacteraceae_ (OTU0017) and
the genus _Thiobacillus_ (OTU0123). Among them, OTU0001 was enriched on all three types of biochar compared with the bulk soil and had a high relative abundance ranging from 7.37 to 27.52%
(with the exception of one Ben–F–particle sample being only 0.68%; Figure 1c). Without fertilization, the relative abundance of OTU0001 on Bam (9.56±2.20%, mean±s.e.m.) was also
significantly lower than on Kao (20.94±2.57%) and Ben (21.74±1.66%). OTU0017 (_Oxalobacteraceae_, class Betaproteobacteria) was enriched only on MEB particles (7.61±1.01% without fertilizer
and 3.13±1.23% with fertilizer) and below the detection limit in the neat biochar. The representative 16S rRNA sequence of OTU0017 was 98% similar to a uncharacterized and unpublished
bacterium isolated from tungsten sand tailings (Genbank accession no. JQ608321; Supplementary Figure 3) and less than 93% similar to the nearest characterized isolate, _Herbaspirillum
massiiliense_ JC206, which was isolated from stool (Lagier et al., 2012). OTU0123 (_Thiobacillus_, class Betaproteobacteria) was abundant on the Kao particles (8.47±3.00%) and only detected
on the Ben particles in the presence of fertilizer (2.14±0.81%). The representative 16S rRNA sequence of OTU0123 was 98% similar to _Thiobacillus thioparus_ strain THI 111 (Genbank accession
no. NR_117864). Our 16S rRNA survey is consistent with a recent study on the surface microbiota of a corncob biochar (Sun et al., 2016), which also observed a strong enrichment of specific
taxa, especially for Proteobacteria (Figure 1b). We further show here that the presence of clay minerals and iron sulfate on the biochar influences the bacterial community composition on the
surface by specifically enriching two Betaproteobacteria (Figures 1b and c). A previous study has seen a similar enrichment of this bacterial class on clay minerals in artificial soils
(Ding et al., 2013). Collectively, this shows that deterministic processes govern the assembly of communities on biochar, which offers the potential to design MEBs that specifically enrich
certain kind of bacteria. GAMMAPROTEOBACTERIUM OTU0001—A REPRESENTATIVE OF A NEW, DIVERGENT BACTERIAL CLADE The prevalence of OTU0001 on all three types of biochar particles and its
low-level taxonomic assignment (that is, class Gammaproteobacteria) led us to further investigate its phylogeny in comparison to related isolates and uncultured organisms (Figure 2). OTU0001
falls into a cluster of uncultured bacteria that were all found in carbonaceous environments. Specifically, the 16S rRNA gene sequence of OTU0001 was identical to an uncultured bacterium
found to be enriched on the surface of a biocathode in a microbial fuel cell (Sun et al., 2012). This biocathode was made from a semicoke, which was obtained from the carbonization of
organic matter below 700 °C, similar to the condition of pyrolysis used here to produce biochar. Other sequences in this cluster were from a cathodic carbon fiber paper (JN802222; Xia et
al., 2012), a submicronic filter of hemodialysis (AM087517; Gomila et al., 2006) and activated carbon filters (DQ646451, JQ237904, JQ926658; Hwang et al., 2006; Liao et al., 2012, 2013).
Overall this shows that OTU0001 represents a new gammaproteobacterial group of uncultured bacteria, whose members are often associated with carbonized materials that can deliver electrons to
cells. This novel group was only distantly related (about 10% 16S rRNA gene sequence divergence) to a cluster of cultured organisms that was defined by _Acidiferrobacter thiooxydans_, which
is an acidophilic iron-oxidizing bacterium (Hallberg et al., 2011). GENOMIC FUNCTION OF THE BIOCHAR-ENRICHED GAMMAPROTEOBACTERIUM To gain insight into the functional properties of the three
biochar/MEB-enriched OTUs, we reconstructed their genomes from metagenomic data (Table 2). A draft genome classified to the class Gammaproteobacteria by Phylosift and with 88.44%
completeness was recovered. The genome termed Gama1 had the highest average sequencing coverage (316 ×) among all recovered genomes and contained a scaffold with a 63 base pairs terminal end
that was identical to the representative 16S rRNA sequence of OTU0001. We therefore assigned the Gama1 genome to the abundant OTU0001. Analysis of the Gama1 genome revealed genes encoding
for the key enzymes involved in carbon fixation cycle (Calvin-Benson-Basham reductive pentose phosphate pathway, CBB), including the RuBisCo large and small subunits and the RuBisCo
activation proteins CbbO, CbbQ and CbbX. We found two types of _RuBisCo_ operon arrangements located in different scaffolds. One gene cluster contained the _RuBisCo_ large and small subunits
followed by _cbbO_ and _cbbQ_ (scaffold ID: Ga0079483_1120) and another one contained the _RuBisCo_ large and small subunits with _cbbX_ located downstream (Ga0079483_1110). These two types
of gene arrangements imply Form IAq and Form IC RuBisCo enzymes, respectively (Badger and Bek, 2008), which were further confirmed by maximum likelihood phylogenetic analysis constructed
for the RbcL (Supplementary Figure 4). The Form IAq RbcL was 94.69% similar to the one from _Lamprocystis purpurea_ DSM 4197 (class _Gammaproteobacteria_), while the Form IC RbcL was 95.49%
similar to _Nitrosococcus halophilus_ Nc4 (class _Gammaproteobacteria_). Both forms of RuBisCo enzymes are adapted to medium to high CO2, but Form IAq is more likely to react with O2 as an
alternative substrate than Form IC (Badger and Bek, 2008). This suggests that Gama1 experiences substantial amounts of CO2 with variable O2 level on the biochar surface. The Gama1 genome
showed versatility with respect to the uptake of nitrogen resources (Figure 3). We identified a gene cluster containing a urease operon with urea transport genes located downstream
(Ga0079483_1050), ammonium, nitrate and nitrite transport genes (Ga0079483_1030 and 1014) as well as assimilatory nitrate reduction genes (Ga0079483_1095). In contrast, ATP-binding cassette
transporters and co-transporters for oligosaccharide and branched-chain amino acid were not recovered in the Gama1 genome (Figure 3), indicating an inability to acquire organic compounds
from the environment. Furthermore, we did not find genes encoding key enzymes for fermentation, such as lactate dehydrogenase, pyruvate decarboxylase and formate dehydrogenase. These
observations suggest that Gama1 has a strictly autotrophic lifestyle. We found a gene cluster in the Gama1 genome (Ga0079483_1017) encoding for a MobB-containing (molybdopterin
oxidoreductase Fe4S4 region) alternative respiratory complex III (AC III), which was first proposed to be involved in iron oxidation in the Zetaproteobacterium _Mariprofundus ferrooxydans_
PV-1 (Singer et al., 2011a, 2013). Gene for the AC III have also recently been found in the Gammaproteobacterium NRL1 (Wang et al., 2015), which shared 88% 16S rRNA gene identity with
OTU0001 (Figure 2). The conceptual iron oxidation pathway by Singer et al. (2013) proposed an outer membrane c-type cytochrome (c-Cyt) and a periplasmic c-Cyt to be involved in electron
transfer. We also observed a gene cluster that consecutively encoded for a c-Cyt biogenesis system, two c-Cyt family proteins with doubled CXXCH heme-binding motifs, three periplasmic
triheme c-Cyt, two porin-like outer membrane proteins and a 2Fe-2S ferredoxin (Ga0079483_1028; Supplementary Figure 5). This operon structure indicates its potential to encode a
porin-cytochrome protein complex for trans-outer-membrane electron transport, similar to what has been described in _Shewanella_ and _Geobacter_ (Lovley et al., 2004; Clarke et al., 2011;
Richardson et al., 2012; Liu et al., 2014). A similar genomic arrangement around the periplasmic triheme c-Cyt gene in this operon (Ga0079483_1028) was also found in an iron-oxidizing
bacterium _Leptothrix cholodnii_ SP-6 (Genbank accession no. NC_010524; Supplementary Figure 5). The porin-cytochrome protein complex might thus function as the outer membrane c-Cyt and
periplasmic c-Cyt in the iron oxidation pathway to conduct electrons from extracellular reduced iron into the electron transport chain (Wang et al., 2015). The Gama1 genome also encodes for
cbb3-type cytochrome c oxidases and a cytochrome bd-I ubiquinol oxidase, which have high affinities for O2 and can thus support growth under microaerophilic conditions (Jünemann, 1997;
Buschmann et al., 2010). Reactive oxygen species, such as superoxide anion radicals, hydrogen peroxide and hydroxyl radicals, are generated during aerobic iron oxidization (Cabiscol et al.,
2010). A series of genes encoding for antioxidants were identified in the Gama1 genome (Supplementary Table S2), including five copies of the cytochrome c peroxidase and two copies of
bacterioferritins. Cytochrome c peroxidases can reduce hydrogen peroxide to water, and have been suggested to be vital during the iron oxidation in _Marinobacter aquaeolei_ (Singer et al.,
2011b; Waite, 2012). Bacterioferritin is an iron-storage protein and is critical for detoxification of harmful iron and oxygen species associated with iron oxidization (Carrondo, 2003).
Although we did not observe homologs to some well-known genes involved in iron oxidation, such as _rus_, _pioABC_, _foxEYZ_ in Gama1 (Bird et al., 2011), based on the putative iron oxidation
pathways found, as well as greater enrichment of Gama1 on the ferrous sulfate treated biochar particles (Kao and Ben) in comparison to untreated biochar particles (Bam, Table 1; Figure 1c),
we propose that the dominant Gama1 oxidizes iron by combining an AC III and porin-cytochrome protein complex (Figure 3). Electrons from iron oxidation could then be transferred to O2, which
generates NADH and ATP to support autotrophic growth. INTERACTION OF GAMA1 WITH IRON ON THE BIOCHAR The genome-based evidence for iron oxidation by Gama1 would predict that the bacterium is
located near iron on the biochar structure. We therefore investigated the physical localization of the Gama1 on the biochar surface using GISH (Ye et al., 2015) and investigated the
elemental distributions and redox states around the bacterium by EDS and EELS. The probe targeting the 16S rRNA gene sequence of OTU0001 hybridized to rod-shaped cells (Figure 4a) through
dense deposition of nanogold particles (Figure 4b). While gold signals were also observed outside of cell structures, they were dispersed and lumpy, which was likely caused by non-specific
binding of nanogold to charged minerals as has been previously reported (Ehrhardt et al., 2009). We also observed pili-like structures on these rod-shaped cells (Figure 4a; arrows),
consistent with the Gama1 genome encoding for pili (Figure 3; Supplementary Information). An EDS spectrum revealed the iron peak in the region around the Gama1 cells (Figure 4c). Other cell
morphotypes without nanogold signal were also found on the same biochar particles (Figures 4d and e), but lacked an iron signal in their vicinity (Figure 4f). This result shows a specific
localization of Gama1 in iron-rich regions of the biochar. To further support the notion of iron oxidization by Gama1, we investigated the iron species that are directly in contact with the
cells using HAADF imaging in STEM combined with EDS to allow SI mapping with a nanometer scale resolution. An EELS was simultaneously conducted to investigate the oxidation state of the iron
(Chen et al., 2009). The bright-field and HAADF images show a nanogold-labeled Gama1 cell next to a particle (Figures 5a and b). We then conducted SI mapping to determine the element
distributions of the cell and the particle (Figure 5a, yellow rectangle). The EDS spectrum shows a strong Au signal (Figure 5c) consistent with the cell being labeled with nanogold as well
as signals for C, N and O (Figure 5d,), as would be expected for cellular matter. The particle has three main phases (Figures 5e–g), which all have similar qualitative elemental composition
of C, O, Mg, Si, S and Fe. This observation indicates that the particle consist of a complex organo-mineral phase. However, the local Fe/O ratios of these three phases are distinct from each
other, with the largest ratio being away from the cell (Figure 5e) and the smallest ratio seen for the region directly in contact with the cell (Figure 5g). This indicates a more reduced
state of Fe species at the distal part of the particle, and a more oxidized state of Fe species in the region directly in contact with the Gama1 cell (Figure 5h). The EELS analysis of the Fe
_2p L_2,3 edge from the proximal area to the Gama1 cell (Figure 5b, inset) showed a small pre-peak in the _L_3 edge (see yellow arrow in inset) and a post-peak in the _L_2 edge in the
region of 710-730 eV, indicating the presence of magnetite (Fe3O4), which contains mixed valence of Fe(II) and Fe(III), and a change in the Fe oxidation state towards γ-Fe2O3 or α-Fe2O3
(Almeida et al., 2014). These observations together further support the notion of active iron oxidation by Gama1, which is taking place on the interface with an iron-containing
organo-mineral particle. The genes putatively involved in the iron oxidation pathway of Gama1 have also been recently proposed to be involved in the extracellular electron uptake from a
cathode in the Gammaproteobacterium NRL1 (Wang et al., 2015). We performed cyclic voltammetry on Bam and the two MEBs (Supplementary Information; Supplementary Figure 6) and found that the
MEBs are considerably more redox active than the Bam. Kao, Ben and Bam are however all able to store and conduct significant amounts of charge. Previous research has found that redox active
magnetic nanoparticles (for example, Fe3O4) can be important for interspecies electron transfer (Aulenta et al., 2013; Klüpfel et al., 2014a, 2014b). Measurements of magnetic hysteresis
loops show that Kao and Ben have a significant concentration of superparamagnetic nanoparticles, whereas Bam does not (Supplementary Information; Supplementary Figures 7 and 8). Nanoscale
examination of the surface of the biochar shows that magnetite particles below 20 nm in size are in direct contact with the Gama1 bacterium (Figure 5i, red arrows). These magnetic iron
nanoparticles could reduce the activation energy for redox reactions to take place and increase the rate of charge transfer (Peng et al., 2013; Yin et al., 2013). We therefore propose that
Gama1 may gain adequate electrons not only through the direct contact with superparamagnetic iron nanoparticles, but also from electrons that are produced in redox reactions in proximal
parts and then conducted through the biochar structure (Joseph et al., 2015). The higher amount of superparamagnetic iron nanoparticles on Kao and Ben and its associated reduction in
activation energy for electron transfer could also explain why the iron-oxidizing Gama1 bacterium is more enriched on these two MEBs (~20% relative abundance; see above) when compared with
Bam (~10%). A PREDICTED MIXOTROPHIC LIFESTYLE OF THE MEB-ENRICHED _OXALOBACTERACEAE_ AND _THIOBACILLUS_ Our metagenomic dataset also contained nearly complete genome sequences assigned to
the family _Oxalobacteraceae_ and the genus _Thiobacillus_, respectively (Table 2). These were termed Oxal1 and Thio1 and assigned to OTU0017 and OTU0123, respectively. Similar to Gama1,
Oxal1 and Thio1 also have complete gene sets for the CBB pathway to carry out carbon fixation (Figure 3). The genomes of Oxal1 and Thio1 harbor genes encoding large and small subunits of
RuBisCo (Oxal1: Ga0079480_116, Thio1: Ga0079481_105). The _RuBisCo_ genes of Oxal1 were classified as Form IAc, whereas Thio1 has two sets of genes for Form IAq and Form II RuBisCo
(Supplementary Figure 4). In general, Form IAc RuBisCo is adapted to low CO2 environment, while Form IAq and II RuBisCo are adapted to medium to high CO2 environment (Badger and Bek, 2008).
In addition, operons encoding carboxysome shell proteins and shell carbonic anhydrase were detected in Oxal1 and Thio1 (Supplementary Figure 4). Carboxysomes are well-known for their role in
encapsulating RuBisCo and carbonic anhydrase, and thereby enhancing carbon fixation by elevating the levels of CO2 in the vicinity of RuBisCo (Yeates et al., 2008). Reconstruction of
central metabolic pathways of Oxal1 and Thio1 revealed essential genes for the glycolysis and tricarboxylic acid cycle (Figure 3). In addition, Oxal1 possesses genes for the Entner–Doudoroff
pathway. Several ATP-binding cassette transporters for branched-chain amino acid, C4-dicarboxylate and oligosaccharide, as well as amino acid and oxalate co-transporters were found in Oxal1
and Thio1 (Figure 3). We also identified genes encoding for enzymes involved in the degradation of complex organic substrates, including aromatic dioxygenases (Oxal1, Ga0079480_102),
protocatechuate-3,4-dioxygenase (Oxal1, Ga0079480_110), toluene monooxygenase (Thio1, Ga0079481_113) and methanesulfonate monooxygenase (Thio1, Ga0079481_113). Acidic, aromatic and phenolic
carbon compounds have previously been found by Nuclear Magnetic Resonance analysis in the two MEBs used here (Rawal et al., 2016) and we found that Ben and Kao contains higher amounts of
dissolved organic carbon than Bam (Table 1). These organic compounds could be utilized by the enzymes above for by Oxal1 and Thio1 for heterotrophic growth on the Ben and Kao surfaces
(Figure 1c). ENERGY GENERATION IN THE MEB-ENRICHED _OXALOBACTERACEAE_ AND _THIOBACILLUS_ Oxal1 and Thio1 were almost exclusively found on the biochar particles treated with ferrous sulfate
(Ben and Kao; Figure 1c), which contained about 10 × more sulfur than Bam (Table 1). X-ray photoelectron spectrometry showed that a substantial proportion of the sulfur is in reduced form in
both fresh and aged Ben and Kao (Supplementary Figure 9). This led us to investigate the sulfur metabolism in these two genomes in more detail. Oxal1 and Thio1 contained gene clusters
encoding for enzymes involved in oxidizing thiolsulfate to sulfate (Friedrich et al., 2005). Specifically, the _sox_ operon in Oxal1 consists of _soxCDYZXAB_ (Ga0079480_117) encoding four
periplasmic proteins, SoxXA, SoxYZ, SoxB and Sox(CD)2 (Figure 3), while the _sox_ operon in Thio1 only contained _soxXYZAB_ (Ga0079481_108). Thio1 also has another two gene clusters
comprised of _soxXA_ (Ga0079481_133) and _soxC_ (Ga0079481_106). Friedrich et al. (2000) reported that the Sox system reconstituted from SoxXA, SoxYZ, SoxB and Sox(CD)2 produced 8 mol of
electrons per mol of thiosulfate, while only 2 mol of electrons were produced with the deletion of Sox(CD)2. The potential absence of _soxCD_ in its genome implies that Thio1 likely utilizes
other oxidization pathways to obtain electrons. We found a _dsrAB_ gene cluster in Thio1 that was similar to the one in _Thiobacillus denitrificans_ ATCC25259 (IMG Genome ID: 637000324, 94%
and 98% similarity for _dsrA_ and _dsrB_, respectively). The _dsrAB_ genes, which encode the siroheme-containing sulfite reductase, has been shown to be involved in the reverse direction
for the dissimilatory oxidation of sulfur in _Thiobacillus denitrificans_ (Trüper, 1994). This indicates that Thio1 may also use dissimilatory oxidation of sulfur compounds to acquire
electrons. The genomes of Oxal1 and Thio1 also encode all necessary enzymes for aerobic respiration (Figure 3). We identified genes encoding four types of terminal oxidases, including aa3-,
bo3- and cbb3-type cytochrome c oxidase, as well as cytochrome bd-I ubiquinol oxidase. Oxal1 contains genes encoding all four types of terminal oxidases, whereas Thio1 harbors genes for
three types of terminal oxidases (that is, no bo3-type cytochrome c oxidase; Supplementary Table 3). It has been shown that both aa3- and bo3-type cytochrome c oxidases are only produced
under O2-rich conditions (Haltia et al., 1988; Cotter et al., 1990), while cbb3-type cytochrome c oxidase and cytochrome bd-I ubiquinol oxidase have high affinities for low levels of O2
(Jünemann, 1997; Buschmann et al., 2010). Hence, the presence of genes encoding both types of terminal oxidases, functioning under either aerobic or microaerobic conditions, suggests a
potential adaptation of Oxal1 and Thio1 to variable O2 levels. Additionally, Thio1 contained a suite of genes for denitrification, including those encoding for nitrate reductase
(_narKKGHJI_, Ga0079481_113), nitrite reductase (_nirK_, Ga0079481_107), nitric oxide reducase (_norZ_, Ga0079481_111) and nitrous oxide reductase (_nosZR_, Ga0079481_113). The capability of
using nitrate/nitrite as terminal electron acceptors may also explain the presence of Thio1 on Kao, which has a higher level of total soluble nitrogen and total N compared to the other two
biochars (Table 1), as well as on Kao and Ben when mono-ammonium phosphate was added (Figure 1c). Additional metabolic properties predicted from the genomes of Oxal1 and Thio1 are presented
in the Supplementary Information. CONCLUSION In contrast to previous 16S rRNA gene-based functional prediction that xenobiotic degradation was enriched on biochar (Sun et al., 2016), we
found that chemolithotrophic and autotrophic metabolisms were characteristic of the abundant bacterial members on the surfaces of biochar and MEBs. A novel, iron-oxidizing Gama1 was found to
dominate all surface communities analyzed here. Iron was provided as part of the production for the MEBs and was also likely adsorbed as nanoparticles from the soil onto the biochar surface
as previously observed (Lin et al., 2012). Furthermore, the electro-conductive properties of biochar (Joseph et al., 2015; Supplementary Information) and the potential ability of direct
electron uptake from solid surfaces by the novel Gammaproteobacterium highlight the possibility that chemolithotrophy could also be driven by redox reactions that occur in any part of the
biochar or the soil environment. This could include anaerobic processes, where bacteria use the biochar surface as electron acceptors, as has been recently shown in laboratory experiments
with _Geobacter sulfurreducens_ (Yu et al., 2015). Temporal fluctuation of redox conditions, for example caused by changes in the soil’s water saturation, might also allow for biochar or MEB
surfaces to be ‘recharged’ through reductive, metabolic processes, similar to what has been recently observed for the redox cycling of humic acids in soil (Klüpfel et al., 2014a, 2014b).
Such anaerobic, biotic processes were however not dominant in our experimental set-up, which was characterized by oxygen-replete conditions. Instead, the genetic capacity for aerobic
respiration was mostly seen in the surface-enriched bacteria. This could allow for aerobic sulfur oxidation to occur in a novel member of the family _Oxalobacteraceae_ (Oxal1) and a
_Thiobacillus_ species (Thio1). The genomes of these two bacteria encode for heterotrophic pathways, which could utilize the complex organic compounds present in MEBs. Their heterotrophic
metabolism would also lead to the enrichment of these two bacteria on the MEBs in comparison to the apparently strict autotrophic Gammaproteobacterium (Figure 1c). However, both the
_Oxalobacteraceae_ and _Thiobacillus_ bacteria, in the same way as the Gammaproteobacterium, may have an extended genetic capacity to CO2 fixation. This indicates that bacteria on the
surface of biochar are indeed limited by available organic substrates. Biochars have both labile and recalcitrant organic compounds (Bruun et al., 2011) and the microbial observations made
here indicate that the metabolizable organic fractions might be largely depleted within the 140 days incubation in our experimental set-up. Long-term exposure of biochar to the soil will
also likely deplete organic compounds suitable to support heterotrophic growth. Biochar and MEB can then support chemolithotrophic processes that provide reductive energy to fuel carbon
fixation. Our results thus provide a microbial mechanism why biochars often have negligible effect on soil respiration (Liu et al., 2016), but instead supports carbon sequestration (Lehmann
et al., 2006; Woolf et al., 2010). Designing biochars and mineral-enriched biochars in the future to improve these kind of microbially mediated redox reactions and carbon sequestration has a
great potential for managing the soil environment. ACCESSION CODES ACCESSIONS GENBANK/EMBL/DDBJ * JQ608321 * NC_010524 * NR_117864 REFERENCES * Abujabhah IS, Bound SA, Doyle R, Bowman JP .
(2016). Effects of biochar and compost amendments on soil physico-chemical properties and the total community within a temperate agricultural soil. _Appl Soil Ecol_ 98: 243–253. Google
Scholar * Almeida TP, Kasama T, Muxworthy AR, Williams W, Nagy L, Hansen TW _et al_. (2014). Visualized effect of oxidation on magnetic recording fidelity in pseudo-single-domain magnetite
particles. _Nat Commun_ 5: 5154. CAS PubMed Google Scholar * Anderson CR, Condron LM, Clough TJ, Fiers M, Stewart A, Hill RA _et al_. (2011). Biochar induced soil microbial community
change: implications for biogeochemical cycling of carbon, nitrogen and phosphorus. _Pedobiologia_ 54: 309–320. CAS Google Scholar * Anderson MJ, Gorley RN, Clarke KR . (2008)
_PERMANOVA+for PRIMER: Guide to Software and Statistical Methods_ 1st edn, PRIMER-E Ltd: Plymouth, UK. Google Scholar * Aulenta F, Rossetti S, Amalfitano S, Majone M, Tandoi V . (2013).
Conductive magnetite nanoparticles accelerate the microbial reductive dechlorination of trichloroethene by promoting interspecies electron transfer processes. _ChemSusChem_ 6: 433–436. CAS
PubMed Google Scholar * Badger MR, Bek EJ . (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. _J Exp Bot_ 59:
1525–1541. CAS PubMed Google Scholar * Bardgett RD, van der Putten WH . (2014). Belowground biodiversity and ecosystem functioning. _Nature_ 515: 505–511. CAS PubMed Google Scholar *
Bird LJ, Bonnefoy V, Newman DK . (2011). Bioenergetic challenges of microbial iron metabolisms. _Trends Microbiol_ 19: 330–340. CAS PubMed Google Scholar * Bruun EW, Hauggaard-Nielsen H,
Ibrahim N, Egsgaard H, Ambus P, Jensen PA _et al_. (2011). Influence of fast pyrolysis temperature on biochar labile fraction and short-term carbon loss in a loamy soil. _Biomass Bioenergy_
35: 1182–1189. CAS Google Scholar * Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H . (2010). The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
_Science_ 329: 327–330. CAS PubMed Google Scholar * Cabiscol E, Tamarit J, Ros J . (2010). Oxidative stress in bacteria and protein damage by reactive oxygen species. _Int Microbiol_ 3:
3–8. Google Scholar * Cao X, Ma L, Liang Y, Gao B, Harris W . (2011). Simultaneous immobilization of lead and atrazine in contaminated soils using dairy-manure biochar. _Environ Sci
Technol_ 45: 4884–4889. CAS PubMed Google Scholar * Carrondo MA . (2003). Ferritins, iron uptake and storage from the bacterioferritin viewpoint. _EMBO J_ 22: 1959–1968. CAS PubMed
PubMed Central Google Scholar * Chen S-Y, Gloter A, Zobelli A, Wang L, Chen C-H, Colliex C . (2009). Electron energy loss spectroscopy and ab initio investigation of iron oxide
nanomaterials grown by a hydrothermal process. _Phys Rev B_ 79: 104103. Google Scholar * Chia C, Munroe P, Joseph S, Lin Y, Lehmann J, Muller D _et al_. (2012). Analytical electron
microscopy of black carbon and microaggregated mineral matter in Amazonian dark Earth. _J Microsc_ 245: 129–139. CAS PubMed Google Scholar * Chia CH, Singh BP, Joseph S, Graber ER, Munroe
P . (2014). Characterization of an enriched biochar. _J Anal Appl Pyrol_ 108: 26–34. CAS Google Scholar * Chia CH, Downie A, Munroe P . (2015) Characteristics of biochar: physical and
structural properties. In: Lehmann J, Joseph S (eds). _Biochar for Environmental Management: Science and Technology_ 2nd edn, Earthscan Books Ltd: London, pp 89–109. Google Scholar * Clarke
TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J _et al_. (2011). Structure of a bacterial cell surface decaheme electron conduit. _Proc Natl Acad Sci USA_ 108: 9384–9389. CAS PubMed
PubMed Central Google Scholar * Cotter PA, Chepuri V, Gennis R, Gunsalus R . (1990). Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in _Escherichia coli_ is regulated by
oxygen, pH, and the fnr gene product. _J Bacteriol_ 172: 6333–6338. CAS PubMed PubMed Central Google Scholar * Darling AE, Jospin G, Lowe E, Matsen FA IV, Bik HM, Eisen JA . (2014).
PhyloSift: phylogenetic analysis of genomes and metagenomes. _PeerJ_ 2: e243. PubMed PubMed Central Google Scholar * Ding G-C, Pronk GJ, Babin D, Heuer H, Heister K, Kögel-Knabner I _et
al_. (2013). Mineral composition and charcoal determine the bacterial community structure in artificial soils. _FEMS Microbiol Ecol_ 86: 15–25. CAS PubMed Google Scholar * Ehrhardt C,
Haymon R, Sievert SM, Holden P . (2009). An improved method for nanogold _in situ_ hybridization visualized with environmental scanning electron microscopy. _J Microsc_ 236: 5–10. CAS
PubMed Google Scholar * Feng Y, Xu Y, Yu Y, Xie Z, Lin X . (2012). Mechanisms of biochar decreasing methane emission from Chinese paddy soils. _Soil Biol Biochem_ 46: 80–88. CAS Google
Scholar * Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. _Proc Natl Acad Sci USA_ 103: 626–631. Article CAS PubMed PubMed Central Google
Scholar * Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S _et al_. (2000). Novel genes coding for lithotrophic sulfur oxidation of _Paracoccus pantotrophus_ GB17.
_J Bacteriol_ 182: 4677–4687. CAS PubMed PubMed Central Google Scholar * Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J . (2005). Prokaryotic sulfur oxidation. _Curr
Opin Microbiol_ 8: 253–259. CAS PubMed Google Scholar * Gomila M, Gasco J, Gil J, Bernabeu R, Inigo V, Lalucat J . (2006). A molecular microbial ecology approach to studying hemodialysis
water and fluid. _Kidney Int_ 70: 1567–1576. CAS PubMed Google Scholar * Hallberg KB, Hedrich S, Johnson DB . (2011). _Acidiferrobacter thiooxydans_, gen. nov. sp. nov.; an acidophilic,
thermo-tolerant, facultatively anaerobic iron-and sulfur-oxidizer of the family _Ectothiorhodospiraceae_. _Extremophiles_ 15: 271–279. CAS PubMed Google Scholar * Haltia T, Puustinen A,
Finel M . (1988). The _Paracoccus denitrificans_ cytochrome aa3 has a third subunit. _Eur J Biochem_ 172: 543–546. CAS PubMed Google Scholar * Harter J, Krause H-M, Schuettler S, Ruser R,
Fromme M, Scholten T _et al_. (2014). Linking N2O emissions from biochar-amended soil to the structure and function of the N-cycling microbial community. _ISME J_ 8: 660–674. CAS PubMed
Google Scholar * Hemkemeyer M, Christensen BT, Martens R, Tebbe CC . (2015). Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial
mineralisation of organic pollutants. _Soil Biol Biochem_ 90: 255–265. CAS Google Scholar * Hwang C, Wu W-M, Gentry T, Carley J, Carroll S, Schadt C _et al_. (2006). Changes in bacterial
community structure correlate with initial operating conditions of a field-scale denitrifying fluidized bed reactor. _Appl Microbiol Biotechnol_ 71: 748–760. Article CAS PubMed Google
Scholar * Jeffery S, Verheijen F, Van Der Velde M, Bastos A . (2011). A quantitative review of the effects of biochar application to soils on crop productivity using meta-analysis. _Agric
Ecosyst Environ_ 144: 175–187. Google Scholar * Joseph S, Husson O, Graber ER, Van Zwieten L, Taherymoosavi S, Thomas T _et al_. (2015). The electrochemical properties of biochars and how
they affect soil redox properties and processes. _Agronomy_ 5: 322–340. CAS Google Scholar * Jünemann S . (1997). Cytochrome bd terminal oxidase. _Biochim Biophys Acta_ 1321: 107–127.
PubMed Google Scholar * Kang DD, Froula J, Egan R, Wang Z . (2015). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. _PeerJ_ 3:
e1165. PubMed PubMed Central Google Scholar * Klüpfel L, Keiluweit M, Kleber M, Sander M . (2014a). Redox properties of plant biomass-derived black carbon (biochar). _Environ Sci Technol_
48: 5601–5611. PubMed Google Scholar * Klüpfel L, Piepenbrock A, Kappler A, Sander M . (2014b). Humic substances as fully regenerable electron acceptors in recurrently anoxic
environments. _Nat Geosci_ 7: 195–200. Google Scholar * Kolton M, Harel YM, Pasternak Z, Graber ER, Elad Y, Cytryn E . (2011). Impact of biochar application to soil on the root-associated
bacterial community structure of fully developed greenhouse pepper plants. _Appl Environ Microbiol_ 77: 4924–4930. CAS PubMed PubMed Central Google Scholar * Kozich JJ, Westcott SL,
Baxter NT, Highlander SK, Schloss PD . (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing
platform. _Appl Environ Microbiol_ 79: 5112–5120. CAS PubMed PubMed Central Google Scholar * Krzywinski M, Altman N . (2014). Points of significance: comparing samples-part II. _Nat
Methods_ 11: 355–356. CAS Google Scholar * Kumar S, Nei M, Dudley J, Tamura K . (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. _Brief
Bioinform_ 9: 299–306. CAS PubMed Google Scholar * Lagier J-C, Gimenez G, Robert C, Raoult D, Fournier P-E . (2012). Non-contiguous finished genome sequence and description of
_Herbaspirillum massiliense_ sp. nov. _Stand Genomic Sci_ 7: 200–209. CAS PubMed PubMed Central Google Scholar * Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA _et
al_. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. _Nat Biotechnol_ 31: 814–821. CAS PubMed PubMed Central Google Scholar *
Langmead B, Salzberg SL . (2012). Fast gapped-read alignment with Bowtie 2. _Nat Methods_ 9: 357–359. CAS PubMed PubMed Central Google Scholar * Larkin MA, Blackshields G, Brown N,
Chenna R, McGettigan PA, McWilliam H _et al_. (2007). Clustal W and Clustal X version 2.0. _Bioinformatics_ 23: 2947–2948. CAS PubMed Google Scholar * Lehmann J, Gaunt J, Rondon M .
(2006). Bio-char sequestration in terrestrial ecosystems–a review. _Mitig Adapt Strategies Glob Chang_ 11: 395–419. Google Scholar * Lehmann J, Rillig MC, Thies J, Masiello CA, Hockaday WC,
Crowley D . (2011). Biochar effects on soil biota–a review. _Soil Biol Biochem_ 43: 1812–1836. CAS Google Scholar * Lehmann J, Joseph S . (2015) _Biochar for Environmental Management:
Science, Technology and Implementation_ 2nd edn. Routledge: London and New York. Google Scholar * Liao X, Chen C, Chang C-H, Wang Z, Zhang X, Xie S . (2012). Heterogeneity of microbial
community structures inside the up-flow biological activated carbon (BAC) filters for the treatment of drinking water. _Biotechnol Bioprocess Eng_ 17: 881–886. CAS Google Scholar * Liao X,
Chen C, Wang Z, Wan R, Chang C-H, Zhang X _et al_. (2013). Changes of biomass and bacterial communities in biological activated carbon filters for drinking water treatment. _Process
Biochem_ 48: 312–316. CAS Google Scholar * Lin Y, Munroe P, Joseph S, Kimber S, Van Zwieten L . (2012). Nanoscale organo-mineral reactions of biochars in ferrosol: an investigation using
microscopy. _Plant Soil_ 357: 369–380. CAS Google Scholar * Liu X, Zheng J, Zhang D, Cheng K, Zhou H, Zhang A _et al_. (2016). Biochar has no effect on soil respiration across Chinese
agricultural soils. _Sci Total Environ_ 554: 259–265. PubMed Google Scholar * Liu Y, Wang Z, Liu J, Levar C, Edwards MJ, Babauta JT _et al_. (2014). A trans‐outer membrane porin‐cytochrome
protein complex for extracellular electron transfer by _Geobacter sulfurreducens_ PCA. _Environ Microbiol Rep_ 6: 776–785. CAS PubMed PubMed Central Google Scholar * Lovley DR, Holmes
DE, Nevin KP . (2004). Dissimilatory fe (iii) and mn (iv) reduction. _Adv Microb Physiol_ 49: 219–286. CAS PubMed Google Scholar * Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E,
Grechkin Y _et al_. (2012). IMG: the integrated microbial genomes database and comparative analysis system. _Nucleic Acids Res_ 40: D115–D122. CAS PubMed Google Scholar * Mills AL .
(2003). Keeping in touch: microbial life on soil particle surfaces. _Adv Agron_ 78: 1–43. Google Scholar * Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA . (2008).
Database indexing for production MegaBLAST searches. _Bioinformatics_ 24: 1757–1764. CAS PubMed PubMed Central Google Scholar * Nielsen S, Minchin T, Kimber S, van Zwieten L, Gilbert J,
Munroe P _et al_. (2014). Comparative analysis of the microbial communities in agricultural soil amended with enhanced biochars or traditional fertilisers. _Agric Ecosyst Environ_ 191:
73–82. Google Scholar * O’Neill B, Grossman J, Tsai M, Gomes J, Lehmann J, Peterson J _et al_. (2009). Bacterial community composition in Brazilian anthrosols and adjacent soils
characterized using culturing and molecular identification. _Microb Ecol_ 58: 23–35. PubMed Google Scholar * Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB _et
al_. (2015). vegan: Community Ecology package. _R package version_ 2: 2–1. Google Scholar * Parks DH, Tyson GW, Hugenholtz P, Beiko RG . (2014). STAMP: statistical analysis of taxonomic and
functional profiles. _Bioinformatics_ 30: 3123–3124. CAS PubMed PubMed Central Google Scholar * Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW . (2015). CheckM: assessing
the quality of microbial genomes recovered from isolates, single cells, and metagenomes. _Genome Res_ 25: 1043–1055. CAS PubMed PubMed Central Google Scholar * Peng X, Yu H, Ai L, Li N,
Wang X . (2013). Time behavior and capacitance analysis of nano-Fe3O4 added microbial fuel cells. _Bioresour Technol_ 144: 689–692. CAS PubMed Google Scholar * Peng Y, Leung HC, Yiu S-M,
Chin FY . (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. _Bioinformatics_ 28: 1420–1428. CAS PubMed Google Scholar *
Pietikäinen J, Kiikkilä O, Fritze H . (2000). Charcoal as a habitat for microbes and its effect on the microbial community of the underlying humus. _Oikos_ 89: 231–242. Google Scholar *
Price MN, Dehal PS, Arkin AP . (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. _PloS One_ 5: e9490. PubMed PubMed Central Google Scholar * Pruesse E,
Peplies J, Glockner FO . (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. _Bioinformatics_ 28: 1823–1829. CAS PubMed PubMed Central Google
Scholar * Quilliam RS, Glanville HC, Wade SC, Jones DL . (2013). Life in the ‘charosphere’–Does biochar in agricultural soil provide a significant habitat for microorganisms? _Soil Biol
Biochem_ 65: 287–293. CAS Google Scholar * Rawal A, Joseph SD, Hook JM, Chia CH, Munroe PR, Donne SW _et al_. (2016). Mineral-biochar composites: molecular structure and porosity. _Environ
Sci Technol_ 50: 7706–7714. CAS PubMed Google Scholar * Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ _et al_. (2012). The ‘porin–cytochrome’model for
microbe‐to‐mineral electron transfer. _Mol Microbiol_ 85: 201–212. CAS PubMed Google Scholar * Rondon MR, Goodman RM, Handelsman J . (1999). The Earth’s bounty: assessing and accessing
soil microbial diversity. _Trends Biotechnol_ 17: 403–409. CAS PubMed Google Scholar * Schmieder R, Edwards R . (2011). Quality control and preprocessing of metagenomic datasets.
_Bioinformatics_ 27: 863–864. CAS PubMed PubMed Central Google Scholar * Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E . (2001). Microbial population structures in soil
particle size fractions of a long-term fertilizer field experiment. _Appl Environ Microbiol_ 67: 4215–4224. CAS PubMed PubMed Central Google Scholar * Singer E, Emerson D, Webb EA,
Barco RA, Kuenen JG, Nelson WC _et al_. (2011a). _Mariprofundus ferrooxydans_ PV-1 the first genome of a marine Fe (II) oxidizing Zetaproteobacterium. _PloS One_ 6: e25386. CAS PubMed
PubMed Central Google Scholar * Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, Pati A _et al_. (2011b). Genomic potential of _Marinobacter aquaeolei_, a biogeochemical
'opportunitroph'. _Appl Environ Microbiol_ 77: 2763–2771. CAS PubMed PubMed Central Google Scholar * Singer E, Heidelberg JF, Dhillon A, Edwards KJ . (2013). Metagenomic
insights into the dominant Fe (II) oxidizing Zetaproteobacteria from an iron mat at Lo'ihi, Hawai'i. _Front Microbiol_ 4: 52. PubMed PubMed Central Google Scholar * Sun D, Meng
J, Xu EG, Chen W . (2016). Microbial community structure and predicted bacterial metabolic functions in biochar pellets aged in soil after 34 months. _Appl Soil Ecol_ 100: 135–143. Google
Scholar * Sun Y, Wei J, Liang P, Huang X . (2012). Microbial community analysis in biocathode microbial fuel cells packed with different materials. _AMB Express_ 2: 21. CAS PubMed PubMed
Central Google Scholar * Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS . (2008). Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues
about Rubisco evolution and structure/function relationships. _J Exp Bot_ 59: 1515–1524. CAS PubMed Google Scholar * Trüper HG . (1994). Reverse siroheme sulfite reductase from
Thiobacillus denitrificans. _Methods Enzymol_ 243: 422–426. Google Scholar * Waite JL . (2012) _Characterization of Cytochrome c Peroxidase of Marinobacter Aquaeolei_. Los Angeles,
California: University of Southern California. Google Scholar * Wang Z, Leary DH, Malanoski AP, Li RW, Hervey WJ, Eddie BJ _et al_. (2015). A previously uncharacterized, nonphotosynthetic
member of the Chromatiaceae is the primary CO2-fixing constituent in a self-regenerating biocathode. _Appl Environ Microbiol_ 81: 699–712. PubMed PubMed Central Google Scholar * Wang J,
Xiong Z, Kuzyakov Y . (2016). Biochar stability in soil: meta-analysis of decomposition and priming effects. _GCB Bioenergy_ 8: 512–523. CAS Google Scholar * Westram R, Bader K, Prüsse E,
Kumar Y, Meier H, Glöckner FO _et al_. (2011). ARB: a software environment for sequence data. In: de Bruijn FJ (ed.) _Handbook of Molecular Microbial Ecology I_. Hoboken, NJ: John Wiley
& Sons, Inc., 399–406. Google Scholar * Woolf D, Amonette JE, Street-Perrott FA, Lehmann J, Joseph S . (2010). Sustainable biochar to mitigate global climate change. _Nat Commun_ 1: 56.
PubMed Google Scholar * Xia X, Sun Y, Liang P, Huang X . (2012). Long-term effect of set potential on biocathodes in microbial fuel cells: electrochemical and phylogenetic
characterization. _Bioresour Technol_ 120: 26–33. CAS PubMed Google Scholar * Yariv S, Cross H . (2001) _Organo-Clay Complexes and Interactions_. Taylor & Francis: New York. Google
Scholar * Ye J, Nielsen S, Joseph S, Thomas T . (2015). High-resolution and specific detection of bacteria on complex surfaces using nanoparticle probes and electron microscopy. _PloS One_
10: e0126404. PubMed PubMed Central Google Scholar * Ye J, Zhang R, Nielsen S, Joseph SD, Huang D, Thomas T . (2016). A combination of biochar-mineral complexes and compost improves soil
bacterial processes, soil quality and plant properties. _Front Microbiol_ 7: 372. PubMed PubMed Central Google Scholar * Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM .
(2008). Protein-based organelles in bacteria: carboxysomes and related microcompartments. _Nat Rev Microbiol_ 6: 681–691. CAS PubMed Google Scholar * Yin Y, Huang G, Tong Y, Liu Y, Zhang
L . (2013). Electricity production and electrochemical impedance modeling of microbial fuel cells under static magnetic field. _J Power Sources_ 237: 58–63. CAS Google Scholar * Yu L, Yuan
Y, Tang J, Wang Y, Zhou S . (2015). Biochar as an electron shuttle for reductive dechlorination of pentachlorophenol by _Geobacter sulfurreducens_. _Sci Rep_ 5: 16221. CAS PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS We acknowledge Dr Simon Hager from the Electron Microscopy Unit and Dr Bill Bin Gong from the Solid State and Elemental Analysis
Unit at UNSW for technical support. We thank Professor Gene Tyson (Australian Centre for Ecogenomics) for advise on the metagenomic sequencing. We also thank Professor Xiaohua Zhang (Ocean
University of China) for kindly providing strain _Catenovulum agarivorans_ YM01. JY would like to thank the support of China Scholarship Council (File ID: 201206230085). This research was
supported by the Australian Research Council (LP120200418) and Renewed Carbon Pty Ltd. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Centre for Marine Bio-Innovation, The University of New
South Wales, Sydney, New South Wales, Australia Jun Ye, Shaun Nielsen & Torsten Thomas * School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, New
South Wales, Australia Jun Ye & Mukan Ji * School of Materials Science and Engineering, The University of New South Wales, Sydney, New South Wales, Australia Stephen D Joseph & Paul
Munroe * Institute of Resource, Ecosystem and Environment of Agriculture, Nanjing Agricultural University, Nanjing, China Stephen D Joseph * Australian Institute of Innovative Materials,
University of Wollongong, Wollongong, New South Wales, Australia David R G Mitchell * School of Environmental and Life Sciences, University of Newcastle, Newcastle, New South Wales,
Australia Scott Donne * Institute for Superconducting and Electronic Materials, University of Wollongong, Wollongong, New South Wales, Australia Joseph Horvat & Jianli Wang * School of
Biological, Earth and Environmental Sciences, The University of New South Wales, Sydney, New South Wales, Australia Torsten Thomas Authors * Jun Ye View author publications You can also
search for this author inPubMed Google Scholar * Stephen D Joseph View author publications You can also search for this author inPubMed Google Scholar * Mukan Ji View author publications You
can also search for this author inPubMed Google Scholar * Shaun Nielsen View author publications You can also search for this author inPubMed Google Scholar * David R G Mitchell View author
publications You can also search for this author inPubMed Google Scholar * Scott Donne View author publications You can also search for this author inPubMed Google Scholar * Joseph Horvat
View author publications You can also search for this author inPubMed Google Scholar * Jianli Wang View author publications You can also search for this author inPubMed Google Scholar * Paul
Munroe View author publications You can also search for this author inPubMed Google Scholar * Torsten Thomas View author publications You can also search for this author inPubMed Google
Scholar CORRESPONDING AUTHOR Correspondence to Torsten Thomas. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary
Information accompanies this paper on The ISME Journal website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOCX 4236 KB) SUPPLEMENTARY TABLES (DOCX 131 KB) RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ye, J., Joseph, S., Ji, M. _et al._ Chemolithotrophic processes in the bacterial communities on the surface of mineral-enriched
biochars. _ISME J_ 11, 1087–1101 (2017). https://doi.org/10.1038/ismej.2016.187 Download citation * Received: 28 June 2016 * Revised: 17 September 2016 * Accepted: 09 December 2016 *
Published: 07 February 2017 * Issue Date: May 2017 * DOI: https://doi.org/10.1038/ismej.2016.187 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative