- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Epstein-Barr virus (EBV) microRNAs (miRNAs) are expressed in EBV-associated tumors and cell lines, but the regulation mechanism of their expression is unclear yet. We investigated
whether the expression of EBV miRNAs is epigenetically regulated in EBV-infected B cell lines. The expression of BART miRNAs was inversely related with the methylation level of the BART
promoter at both steady-state and following 5-aza-2'-deoxycytidine treatment of the cells. The expression of BHRF1 miRNAs also became detectable with the demethylation of Cp/Wp in
latency I EBV-infected cell lines. Furthermore, _in vitro_ methylation of the BART and Cp promoters reduced the promoter-driven transactivation. In contrast, tricostatin A had little effect
on the expression of EBV miRNA expression as well as on the BART and Cp/Wp promoters. Our results suggest that promoter methylation, but not histone acetylation, plays a role in regulation
of the EBV miRNA expression in EBV-infected B cell lines. SIMILAR CONTENT BEING VIEWED BY OTHERS CROSS-SPECIES CHROMATIN INTERACTIONS DRIVE TRANSCRIPTIONAL REWIRING IN EPSTEIN–BARR
VIRUS–POSITIVE GASTRIC ADENOCARCINOMA Article 27 July 2020 TARGETING THE SIGNALING IN EPSTEIN–BARR VIRUS-ASSOCIATED DISEASES: MECHANISM, REGULATION, AND CLINICAL STUDY Article Open access 12
January 2021 IDENTIFICATION AND CHARACTERIZATION OF A NOVEL EPSTEIN-BARR VIRUS-ENCODED CIRCULAR RNA FROM _LMP-2_ GENE Article Open access 13 July 2021 INTRODUCTION Epstein-Barr virus (EBV)
is a ubiquitous human herpesvirus implicated in several lymphomas and carcinomas (Young and Rickinson, 2004). Three different latency patterns have been demonstrated depending on the use of
distinct latency promoters. In latency III, long (up to 120 kb) primary transcripts synthesized from activated _Bam_HI C promoter (Cp) or _Bam_HI W promoter (Wp) are alternatively spliced to
produce mature transcripts encoding six different EBV nuclear antigen (EBNA) proteins (EBNA-1, -2, -3A, -3B, -3C, and -LP) (Abbot et al., 1990). In latency I and II, _Bam_HI Q promoter (Qp)
is activated to produce EBNA1, while both Wp and Cp become silent due to DNA methylation (Li and Minarovits, 2003; Tao and Robertson, 2003). In addition, EBV-encoded RNAs (EBERs) and
_Bam_HI A rightward transcripts (BARTs) are expressed in all latently-infected cells (Middeldorp et al., 2003; van Beek et al., 2003). BART transcripts were originally identified in the nude
mouse-passaged nasopharyngeal carcinoma (NPC) cell line C15 (Hitt et al., 1989; Gilligan et al., 1990) and also known as the complementary strand transcripts (CSTs) or BARF0 RNAs (Gilligan
et al., 1990; Smith, 2001). BART transcripts are expressed at particularly high levels in NPC but also expressed in EBV infected peripheral blood B cells (Chen et al., 1992; Brooks et al.,
1993). EBV encodes two groups of viral microRNAs (miRNAs): BHRF1 miRNAs and BART miRNAs. Expression of the EBV miRNAs has been observed not only in EBV-infected cell lines (Pfeffer et al.,
2004; Cai et al., 2006; Grundhoff et al., 2006) but also in EBV-associated tumors (Cai et al., 2006; Kim et al., 2007; Jun et al., 2008). The expression pattern of BHRF1 miRNAs in certain
cells is related to the EBV latencies in those cells (Pfeffer et al., 2004; Cai et al., 2006; Grundhoff et al., 2006). BHRF1 miRNAs sequences are located within introns of the BHRF1 gene
that lies downstream of Cp/Wp and upstream of Qp (Pfeffer et al., 2004; Grundhoff et al., 2006) and transcribed as part of the long EBNA transcripts during latency III (Cai et al., 2006). In
latency I and II, the long transcripts encompassing EBNAs and BHRF1 miRNAs cannot be produced as Cp/Wp are silenced due to DNA methylation (Li and Minarovits, 2003; Tao and Robertson,
2003). Unlike the latency III restricted BHRF1 miRNAs expression, BART miRNAs are expressed in all three latencies (Pfeffer et al., 2004; Cai et al., 2006). The expression level of BART
miRNAs correlates with the expression of BART transcripts in EBV-infected cell lines (Cai et al., 2006; Edwards et al., 2008; Amoroso et al., 2011), supporting the notion that BART miRNAs
are processed from the BART transcripts. Recently, Pratt _et al._ (Pratt et al., 2009) reported that BART miRNAs are expressed at 25-50-fold different levels in various EBV-infected cell
lines. They found that the steady-state levels of BART miRNAs in EBV-infected cell lines did not correlate with the number of viral templates per cell and were not elevated by lytic
induction (Pratt et al., 2009). Thus far, it is unclear why the level of BART miRNAs expression varies greatly in different cells. miRNA expression can be regulated transcriptionally and
post-transcriptionally. Several transcription factors have been shown to activate or repress the transcription of primary miRNAs (Chen et al., 2005; O'Donnell et al., 2005; Rao et al.,
2006; He et al., 2007). miRNA processing can be modulated by RNA-binding proteins. For example, Lin-28 and Lin-28B inhibit let-7 maturation by inhibiting primary miRNA or precursor miRNA
cleavage (Heo et al., 2008; Viswanathan et al., 2008), while HnRNP A1 enhances processing of miR-18a (Guil and Cáceres, 2007; Michlewski et al., 2008). Expression of miRNAs is also known to
be regulated by epigenetic mechanisms, such as promoter methylation (Bueno et al., 2008; Toyota et al., 2008) or histone acetylation (Scott et al., 2006; Grady et al., 2008; Saito et al.,
2009). In this study, we investigated if the expression of BART miRNAs is epigenetically regulated in EBV-positive B cell lines. RESULTS INVERSE CORRELATION BETWEEN BART MIRNAS EXPRESSION
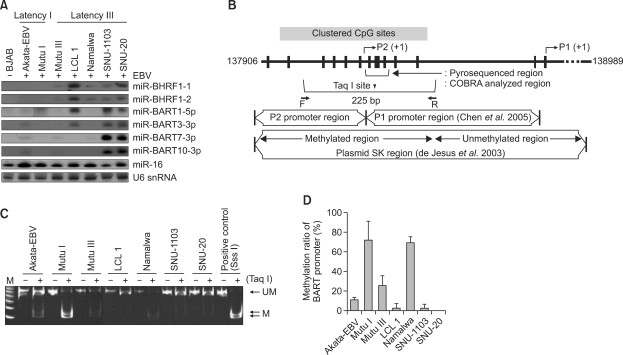
AND METHYLATION STATUS OF THE PROMOTER The expression of EBV miRNAs in two latency I and five latency III EBV-infected B cell lines were analyzed by Northern blot. As expected, BHRF1 miRNAs
were readily detectable in latency III cells but were not detected in any of the latency I cells (Figure 1A). Although BART miRNAs are known to be expressed in all the EBV infected cells,
their expression level varied greatly in different cells. In general, lymphoblastoid cell lines (LCL) (LCL1, SNU-1103 and SNU-20) expressed BART miRNAs at markedly higher level than
Burkitt's lymphoma cell lines (Akata-EBV, Mutu I, Mutu III and Namalwa). LCL1 did not express miR-BART7-3p and miR-BART10-3p as it is infected with B95-8 EBV strain which has a large
deletion in these regions (Wei et al., 1994). To test if the expression level of BART miRNAs was related with epigenetic status of the BART promoter, methylation level of the BART promoter
was analyzed by COBRA using a primer set capable of amplifying the sequence encompassing the clustered CpG sites in this region (Figure 1B). BART promoters of LCL1, SNU-1103, and SNU-20 were
unmethylated for the most part, while those of Akata-EBV, Mutu I, Mutu III, and Namalwa were methylated at variable degrees (Figure 1C). To confirm the results of COBRA, pyrosequencing was
carried out for the four consecutive CpG sites (Figure 1B) within the BART promoter of the cells. Methylation ratios of the four CpG sites in the BART promoter region were not appreciably
different from each other within each cell line (Supplemental Data Table S1). However, different cells showed differential methylation ratios in their BART promoters (Supplemental Data Table
S1; Figure 1D). LCLs that showed a high level miRNA expression such as LCL1, SNU-1103, and SNU-20 exhibited relatively low BART promoter methylation, while Mutu I and Namalwa which had
lowest BART miRNAs expression displayed the highest promoter methylation ratios among the tested cell lines (Figure 1A vs. Figure 1D). Akata-EBV and Mutu III showed intermediate levels of
BART promoter methylation and BART miRNAs expression. UPREGULATION OF BART MIRNAS EXPRESSION FOLLOWING DEMETHYLATION OF THE BART PROMOTER USING 5-AZA-CD If hypermethylation of the CpG-rich
area was one of the cause of transcriptional repression of the BART miRNAs, demethylation of the CpGs should upregulate their expression. To check whether BART miRNAs expression could be
induced by BART promoter demethylation, cells were treated with 5-aza-2'-deoxycytidine (5-aza-Cd) for 72 h. BART promoter methylation decreased significantly following 0.5-5 µM 5-aza-Cd
treatment of Mutu I cells compared to the untreated control (Figure 2A, Supplemental Data Table S2). This coincided with a large induction of BART miRNAs expression in this cell line
(Figure 2B). BART promoter methylation decreased (Figure 2A) and BART miRNAs expression increased noticeably (Figure 2B) only when these cells were treated with higher concentrations of
5-aza-Cd than 5 µM. In Akata-EBV, which showed relatively low basal BART promoter methylation, 5-aza-Cd treatment reduced methylation ratio of the promoter only slightly and BART miRNAs
expression was induced nominally. 5-Aza-Cd treatment of the cells which have hypomethylated promoters such as LCL1, SNU-1103, and SNU-20 failed to alter the levels of BART miRNAs expression
as well as the promoter methylation (data not shown). To investigate whether the expression of EBV miRNAs could also be regulated by histone acetylation, BART and BHRF1 miRNAs expression was
analyzed for latency I cells treated with trichostatin A (TSA) alone or in combination with 5-aza-Cd. TSA cotreatment with 5-aza-Cd or TSA alone had no effect on the BART and BHRF1 miRNAs
expression (Figure 3A) as well as on the promoter methylation (Figure 3B). Not unexpectedly, the expression of BHRF1 miRNAs was detected in latency I Akata-EBV and Mutu I cells treated with
5-aza-Cd (Figure 3A). As BHRF1 miRNAs are reported to be expressed in only latency III, we checked whether treatment with 5-aza-Cd caused activation of Cp/Wp in these latency I cells. Cp and
Wp transcripts were detected in Akata-EBV and Mutu I cells following treatment with 5-aza-Cd but not with TSA (Figure 3C). A transcript initiating at Qp was detected in these cells
regardless of the treatment. Expression of EBNA-2 was also observed by RT-PCR and Western blot in these cells after treated with 5-aza-Cd (Figure 3D), confirming the activation of latency
III promoters. We did not noticed significant alteration in the size and shape of Mutu I and Akata-EBV cell following 5-aza-Cd treatment using bright-field microscopy. To make sure TSA was
functioning properly, cell cycle was accessed by flow cytometric analysis following TSA-treatment of the Akata-EBV and Mutu I cells. Cell cycle was inhibited when these two cell lines were
treated with increasing concentrations of TSA for 24 h (Supplemental Data Figure S1). However, the expression of BART miRNAs was not induced in the same cells following up to 100 nM TSA
treatment (Figure 3E). TSA had no effect on the expression of BART and BHRF1 miRNAs in latency III cell lines, either (Figure 4). INDUCTION OF EBV LYTIC CYCLE BY 5-AZA-CD AND TSA TREATMENT
The effect of 5-aza-Cd and TSA on the induction of EBV lytic cycle was analyzed by detecting the expression of BMRF1 gene product (early antigen diffuse component; EA-D) (Figure 5).
Akata-EBV, Mutu I, Mutu III, and Namalwa cells were treated with 5-aza-Cd (5 or 50 µM) for 72 or TSA (100 nM) for 24 h. 5-Aza-Cd and TSA treatment caused EA-D expression in small portions of
Akata-EBV (5-aza-Cd, 27%; TSA, 7%) and Mutu I (5-aza-Cd, 41%; TSA, 12%) cells but not in Mutu III and Namalwa (Figure 5). 5-Aza-Cd induced EA-D expression seemed to coincide with the
enhanced BART miRNA expression (Figure 5 vs. Figure 2B). However, there were no parallels between the extent of lytic cycle induction and the extent of upregulated miRNA expression by TSA
(Figure 5 vs. Figure 3E). REPRESSION OF BART PROMOTER AND CP ACTIVITY VIA _IN VITRO_ METHYLATION Cells exhibiting hypermethylated promoter region showed enhanced expression of BART miRNAs
when they were treated with 5-aza-Cd. To determine whether DNA methylation directly activates the BART promoter, _in vitro_ methylation assays were performed. The CpG residues within the
BART promoter was methylated _in vitro_ using the CpG-specific SssI methylase, which was confirmed by digestion with the methylation sensitive restriction endonuclease _Hpa_II (Figure 6A).
Each of the unmethylated and methylated BART promoter fragment was ligated with pGL3-basic vector and transfected into AGS cells. Forty-eight hours after transfection, luciferase activities
were measured to determine the BART promoter activity. _In vitro_ methylation of the CpG residues within the BART promoter repressed the basal BART promoter activity by 77% (Figure 6B). As
previously reported (Robertson et al., 1995; Robertson and Ambinder, 1997), Cp methylation also significantly reduced the basal and EBNA2-induced Cp activation (Figure 6C). DISCUSSION In our
experiments, BL cell lines expressed BART miRNAs at lower levels than LCLs. In general, BL biopsies and BL cell lines show a high level EBV DNA methylation, whereas LCLs show unmethylated
or hypomethylated EBV DNA (Minarovits et al., 1991). We found that BART promoters present in BL cell lines were more heavily methylated than those in LCLs. In B cell lines, 5-aza-Cd
treatment reduced BART promoter methylation and induced BART miRNAs expression. Furthermore, in vitro methylation of the CpG residues within the BART promoter significantly reduced BART
promoter activity, supporting direct link between BART promoter methylation and BART miRNAs expression. Epigenetic status of the EBV BART promoter region has been analyzed by several
investigators previously (Smith et al., 2000; de Jesus et al., 2003; Chen et al., 2005; Al-Mozaini et al., 2009). One group reported that BART promoter activity was found in about 1 kb
region encompassing just upstream of the transcription start site to a distance downstream of exon I of the BART gene (de Jesus et al., 2003; Al-Mozaini et al., 2009). The upstream sequences
of the BART promoter including the CpG residues (Figure 1B) was shown to be methylated in DNA from C15 NPC tissue and argued that this region was not essential for BART transcripts
expression (de Jesus et al., 2003). However, their data showed that deletion of the plasmids containing this upstream sequences, including the CpG residues that we analyzed in this study,
resulted in enhanced BART promoter activity (de Jesus et al., 2003). This supports that the upstream sequences including the CpG residues indeed down-regulated the BART promoter, possibly by
DNA methylation as we suggested in this paper. Another group mapped two different TATA-less promoter regions (P1 and P2) for BART transcripts (Chen et al., 2005). Both P1 and P2 are located
upstream of exon I. P1 appears to be the dominant start site for BARTs expression in Akata cell line and NPC tumor tissues. The region encompassing both P1 and P2 promoters is
hypomethylated in C15 NPC-tumor-associated virus. The BART promoter region analyzed in our experiments expands from 100 bp upstream of the P2 transcription start site to 259 bp upstream of
the P1 transcription start site (Figure 1). In some cells such as Akata-EBV, BART miRNAs expression was not significant even though the BART promoter methylation was relatively low. Thus,
the expression of BART miRNAs may also be regulated by other mechanisms (Palii et al., 2008; Dickerson et al., 2009) in addition to the BART promoter methylation in these cells. For example,
P1 promoter was downregulated by IRFs, while P2 promoters were positively regulated by C-Myc and C/EBP family members (Chen et al., 2005). Differential expression level of these
transcription factors in different cell lines could also affect BART miRNAs expression. Treatment with 5-aza-Cd may have induced those transcription factors (Palii et al., 2008) to affect
EBV miRNAs expression in addition to directly affected the methylation status of EBV miRNA promoters. miRNA processing and degradation might also be affected by 5-aza-Cd treatment (Saito et
al., 2006; Chuang and Jones, 2007). We found that Cp/Wp of latency I cells became activated and BHRF1 miRNAs were expressed after 5-aza-Cd treatment. Our results corroborate that BHRF1
miRNAs are primarily processed from the long EBNA transcripts produced from Cp/Wp in latency III (Cai et al., 2006; Xing and Kieff, 2007; Amoroso et al., 2011). Latency I cells treated with
TSA showed a high level of acetylated histone H4 at Cp, but the transcripts initiated at Cp were detected at very low levels (Fejer et al., 2008). This may explain why we could not detect
the expression of BHRF1 miRNAs in latency I cells following TSA treatment. It is controversial whether BART miRNA expression is affected by EBV lytic induction. Pratt _et al._ (Pratt et al.,
2009) showed that the level of BART miRNAs were not changed by lytic induction of EBV infected Akata and B95-8 cells using IgG and TPA treatment, respectively. Xing and Kieff (Xing and
Kieff, 2007) also reported that miR-BART1 and miR- BART-2 expression was not affected following induction of EBV replication in Akata cells. However Cai _et al._ (Cai et al., 2006) argued
that the expression BART miRNAs was increased significantly in Mutu I cells lytically induced with TPA and n-butyrated. In this study, treatment with 5-aza-Cd or TSA caused EBV lytic cycle
in latency I cells as we previously reported (Jung et al., 2007; Seo et al., 2008). Even though, the extent of EBV lytic induction by 5-aza-Cd seem to correlate with the increase of miRNA
expression, the extent of EBV lytic induction by TSA did not parallel the extent of upregulated miRNA expression. There are two plausible reasons for these seemingly contradictory results.
First, achieving a certain level of EBV lytic induction may be important to upregulate BART miRNAs. Second, not the lytic induction per se, but some factor(s) affected by 5-aza-Cd or TSA
treatment may regulate BART miRNA transcription, procession, or degradation. Further studies are required to clarify these possibilities. Taken together, our data suggest that promoter
methylation, but not histone acetylation, plays a partial role in the regulation of EBV BART and BHRF1 miRNAs expression in EBV infected B cells. METHODS CELL LINES AND CULTURE MEDIUM
EBV-positive (Akata-EBV, Mutu I, Mutu III, Namalwa, LCL 1, SNU-1103, and SNU-20) and EBV-negative (BJAB and AGS) cell lines were used. Akata was provided by Dr. K. Takada (Hokkaido
University, Japan). Mutu I, Mutu III, Namalwa, LCL1, and BJAB were provided by Dr. R. Longnecker (Northwestern University). AGS, SNU-20, and SNU-1103 were obtained from Korea Cell Line Bank
(Seoul, Korea). The cells were maintained at 37℃ in RPMI-1640 plus 10% fetal bovine serum (FBS; Gibco, Grand Island, NY), antibiotics (penicillin 100 units/ml and streptomycin 100 µg/ml,
Gibco), and Fungizone (100 µg/ml, Gibco). To assess the effect of epigenetic modulation on miRNA expression, cells (5 × 105/ml) were treated with increasing concentrations of
5-aza-2'-deoxycytidine (5-aza-Cd) and/or TSA for 24 or 72 h. NORTHERN BLOT OF EBV MIRNAS RNA extraction was conducted with RNAzol™ B reagent (Tel-Test, Friendswood, TX) according to the
manufacturer's instruction. Equal amounts of total RNA (30 µg) were resolved in a 15% polyacrylamide-8 M urea gel and transferred to a Zeta-Probe Blotting membrane (Bio-Rad
Laboratories, Hercules, CA). An oligonucleotide complementary to each mature EBV miRNA (Kim et al., 2007; Jun et al., 2008) was end-labeled with [γ-32P] ATP and T4 kinase. Pre-hybridization
and hybridization were carried out using ExpressHyb Hybridization Solution (BD Biosciences Clontech, Palo Alto, CA). Quality and quantity of the loaded RNA was assessed by reprobing the blot
for U6 snRNA using a [γ-32P]-labeled oligodeoxynucleotide probe (5'-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3'). ANALYSIS OF EBV LATENT PROMOTER USAGE First-strand cDNA synthesis was
performed using 1 µg total RNA (Lee et al., 2008) and a 3'-primer specific for each transcript initiating at Cp, Wp, or Qp EBV promoter. The cDNA samples were then subjected to 30
cycles of PCR using appropriate primer pairs for the transcripts initiating at Cp, Wp, or Qp (Sugiura et al., 1996). Each cycle consisted of denaturation for 30 s at 94℃, annealing for 30 s
at 59℃, and extension for 30 s at 72℃. The PCR products were then electrophoresed in a 1.5% agarose gel, blotted onto a Hybond N+ nylon membrane (Amersham, Buckinghamshire, UK), and analyzed
with an enhanced chemiluminescence 3'-oligolabeling and detection system (Amersham). To detect EBNA2 gene expression, Western blot and RT-PCR were performed as described elsewhere (Oh
et al., 2007; Kubota et al., 2008). DNA EXTRACTION AND BISULFITE TREATMENT DNA was extracted from the cells by standard phenolchloroform extraction. Bisulfite treatment was performed for 1
µg genomic DNA using an EZ DNA™ methylation kit (Zymo Research, Orange, CA). Bisulfite-treated DNA was used for Combined Bisulfite Restriction Analysis (COBRA) and pyrosequencing. COMBINED
BISULFITE RESTRICTION ANALYSIS (COBRA) PCR primers were designed to amplify a series of CpG dinucleotides within the reported BART promoter region (Smith et al., 1993; de Jesus et al., 2003;
Chen et al., 2005): forward primer 5'-AGGGTAAGGTTATAATTGTAATTTT-3' and reverse primer 5'-(Biotin) ATCTATTTATACATTTTTCT CAAATCTAAT-3'. The PCR product (225 bp) was
digested with TaqI (New England Biolabs, Ipswich, MA), separated in a 10% acrylamide gel, and visualized by ethidium bromide staining. PYROSEQUENCING A separate PCR was carried out using the
same primers and templates as for COBRA and the PCR product was used for pyrosequencing. Pyrosequencing was performed using the PSQ96MA system (Biotage AB, Uppsala, Sweden) and PyroGold
reagents (Biotage AB). The primer used for pyrosequencing was 5'-TCCCCGGGAGTGTATC-3'. PLASMIDS pGL3-BART promoter was constructed by PCR amplification of the BART promoter with the
following primers: 5'-GGGGTACCGTAGCTACGGCCAAGGGCAG-3' and 5'-CCCAAGCTTGCAGCTTGAAAAATGGCAAC-3' (Robertson and Ambinder, 1997; Chen et al., 2005) The PCR product was
digested with _Kpn_I and _Hind_III, and then cloned into the corresponding sites of pGL3-basic vector (Promega, Madison, WI). pSG5-EBNA2 (wild type) was kindly provided by Dr. Elliott Kieff
(Harvard Medical School). IMMUNOFLUORESCENCE ASSAY Mutu I and Mutu III were seeded in a 10-well slide. The cells were fixed in 100% ice-methanol for 5 min at -20℃. For immunofluorescence
staining, EA-D (mouse monoclonal antibody (Novocastra, Newcastle upon Tyne, UK, 1:500) was used. Neclei were stained with Prolong Gold Anti-fade Reagent (Molecular probe, Invitrogen,
Carlsbad, CA) containing 4', 6-diamino-2-phenylinodole (DAPI). The fluorescence signal was visualized using a Fluorescence Attached Microscope (AX70, TR-62A02, Olympus, Tokyo, JAPAN).
_IN VITRO_ METHYLATION, TRANSFECTION, AND LUCIFERASE ASSAY The BART promoter fragment was excised from pGL3-BART promoter using _Kpn_I and _Hind_III, and gel purified with the QIAquick gel
extraction kit (QIAGEN, Valencia, CA). The BART promoter fragment was then incubated overnight with SssI methylase (New England Biolabs). The methylation efficiency was monitored by _Hpa_II
(New England Biolabs) digestion and gel electrophoresis. Each of the unmethylated and methylated BART promoter fragment was ligated into the pGL3-basic vector and transfected along with
_Renilla_ luciferase reporters (pRL-TK) into AGS cells using Lipofectamine 2000 transfection reagents (Invitrogen, Carlsbad, CA). AGS cells were used for this analysis as they are widely
used for EBV research and easily transfected. In addition, AGS cells infected with recombinant EBV were shown to express BART miRNAs at high levels (Kim et al., 2007). Forty-eight hours
after transfection, luciferase activities were measured by Dual-Glo Luciferase assay system (Promega). Each transfection was performed in triplicate, and experiments were repeated three
times. Luciferase activity was normalized for transfection efficiency by pRL-TK _Renilla_ luciferase activity. ABBREVIATIONS * 5-aza-Cd: 5-aza-2'-deoxycytidine * BARTs: _Bam_HI A
rightward transcripts * Cp: _Bam_HI C promoter * CSTs: complementary strand transcripts * EBERs: EBV-encoded RNAs * EBNA: EBV nuclear antigen * EBV: Epstein-Barr virus * LCL: lymphoblastoid
cell lines * miRNA: microRNAs * NPC: nasopharyngeal carcinoma * Qp: BamHI Q promoter * TSA: trichostatin A * Wp: _Bam_HI W promoter REFERENCES * Abbot SD, Rowe M, Cadwallader K, Ricksten A,
Gordon J, Wang F, Rymo L, Rickinson AB . Epstein-Barr virus nuclear antigen 2 induces expression of the virus-encoded latent membrane protein . _J Virol_ 1990 ; 64 : 2126 - 2134 CAS PubMed
PubMed Central Google Scholar * Al-Mozaini M, Bodelon G, Karstegl CE, Jin B, Al-Ahdal M, Farrell PJ . Epstein-Barr virus BART gene expression . _J Gen Virol_ 2009 ; 90 : 307 - 316
Article CAS PubMed Google Scholar * Ambros V . The functions of animal microRNAs . _Nature_ 2004 ; 431 : 350 - 355 Article CAS PubMed Google Scholar * Amoroso R, Fitzsimmons L,
Thomas WA, Kelly GL, Rowe M, Bell AI . Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation . _J Virol_ 2011 ; 85 : 996 - 1010 Article
CAS PubMed Google Scholar * Brooks LA, Lear AL, Young LS, Rickinson AB . Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency . _J
Virol_ 1993 ; 67 : 3182 - 3190 CAS PubMed PubMed Central Google Scholar * Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J,
Malumbres M . Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression . _Cancer Cell_ 2008 ; 13 : 496 - 506 Article CAS PubMed Google Scholar *
Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR . Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed . _PLoS Pathog_
2006 ; 2 : e23 - Article PubMed PubMed Central Google Scholar * Chen H, Huang J, Wu FY, Liao G, Hutt-Fletcher L, Hayward SD . Regulation of expression of the Epstein-Barr virus BamHI-A
rightward transcripts . _J Virol_ 2005 ; 79 : 1724 - 1733 Article CAS PubMed PubMed Central Google Scholar * Chen HL, Lung MM, Sham JS, Choy DT, Griffin BE, Ng MH . Transcription of
BamHI-A region of the EBV genome in NPC tissues and B cells . _Virology_ 1992 ; 191 : 193 - 201 Article CAS PubMed Google Scholar * Chuang JC, Jones PA . Epigenetics and microRNAs .
_Pediatr Res_ 2007 ; 61 : 24R - 29R Article CAS PubMed Google Scholar * de Jesus O, Smith PR, Spender LC, Elgueta Karstegl C, Niller HH, Huang D, Farrell PJ . Updated Epstein-Barr virus
(EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV . _J Gen Virol_ 2003 ; 84 : 1443 - 1450 Article CAS PubMed Google Scholar * Dickerson SJ, Xing Y,
Robinson AR, Seaman WT, Gruffat H, Kenney SC . Methylation-dependent binding of the epstein-barr virus BZLF1 protein to viral promoters . _PLoS Pathog_ 2009 ; 5 : e1000356 - Article PubMed
PubMed Central Google Scholar * Edwards RH, Marquitz AR, Raab-Traub N . Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing . _J Virol_ 2008 ; 82 : 9094
- 9106 Article CAS PubMed PubMed Central Google Scholar * Fejer G, Koroknai A, Banati F, Gyory I, Salamon D, Wolf H, Niller HH, Minarovits J . Latency type-specific distribution of
epigenetic marks at the alternative promoters Cp and Qp of Epstein-Barr virus . _J Gen Virol_ 2008 ; 89 : 1364 - 1370 Article CAS PubMed Google Scholar * Gilligan K, Sato H, Rajadurai P,
Busson P, Young L, Rickinson A, Tursz T, Raab-Traub N . Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma . _J Virol_ 1990 ; 64
: 4948 - 4956 CAS PubMed PubMed Central Google Scholar * Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM, Kroh EM, Allen A,
Fritz BR, Markowitz SD, Tewari M . Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer . _Oncogene_ 2008 ; 27 : 3880 - 3888 Article CAS
PubMed Google Scholar * Grundhoff A, Sullivan CS, Ganem D . A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses . _RNA_
2006 ; 12 : 733 - 750 Article CAS PubMed PubMed Central Google Scholar * Guil S, Cáceres JF . The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a .
_Nat Struct Mol Biol_ 2007 ; 14 : 591 - 596 Article CAS PubMed Google Scholar * He L, He X, Lowe SW, Hannon GJ . microRNAs join the p53 network--another piece in the tumour-suppression
puzzle . _Nat Rev Cancer_ 2007 ; 7 : 819 - 822 Article CAS PubMed PubMed Central Google Scholar * Heo I, Joo C, Cho J, Ha M, Han J, Kim VN . Lin28 mediates the terminal uridylation of
let-7 precursor MicroRNA . _Mol Cell_ 2008 ; 32 : 276 - 284 Article CAS PubMed Google Scholar * Hitt MM, Allday MJ, Hara T, Karran L, Jones MD, Busson P, Tursz T, Ernberg I, Griffin BE .
EBV gene expression in an NPC-related tumour . _EMBO J_ 1989 ; 8 : 2639 - 2651 Article CAS PubMed PubMed Central Google Scholar * Jun SM, Hong YS, Seo JS, Ko YH, Yang CW, Lee SK .
Viral microRNA profile in Epstein-Barr virus-associated peripheral T cell lymphoma . _Br J Haematol_ 2008 ; 142 : 320 - 323 Article CAS PubMed Google Scholar * Jung EJ, Lee YM, Lee BL,
Chang MS, Kim WH . Lytic induction and apoptosis of Epstein-Barr virus-associated gastric cancer cell line with epigenetic modifiers and ganciclovir . _Cancer Lett_ 2007 ; 247 : 77 - 83
Article CAS PubMed Google Scholar * Kim DN, Chae HS, Oh ST, Kang JH, Park CH, Park WS, Takada K, Lee JM, Lee WK, Lee SK . Expression of viral microRNAs in Epstein-Barr virus-associated
gastric carcinoma . _J Virol_ 2007 ; 81 : 1033 - 1036 Article CAS PubMed Google Scholar * Kubota N, Wada K, Ito Y, Shimoyama Y, Nakamura S, Nishiyama Y, Kimura H . One-step multiplex
real-time PCR assay to analyse the latency patterns of Epstein-Barr virus infection . _J Virol Methods_ 2008 ; 147 : 26 - 36 Article CAS PubMed Google Scholar * Lee W, Choi HI, Kim MJ,
Park SY . Depletion of mitochondrial DNA up-regulates the expression of MDR1 gene via an increase in mRNA stability . _Exp Mol Med_ 2008 ; 40 : 109 - 117 Article CAS PubMed PubMed Central
Google Scholar * Lee WK, Kim SM, Sim YS, Cho SG, Park SH, Kim CW, Park JG . B-lymphoblastoid cell lines from cancer patients . _In Vitro Cell Dev Biol Anim_ 1998 ; 34 : 97 - 100 Article
CAS PubMed Google Scholar * Li H, Minarovits J . Host cell-dependent expression of latent Epstein-Barr virus genomes: regulation by DNA methylation . _Adv Cancer Res_ 2003 ; 89 : 133 -
156 Article CAS PubMed Google Scholar * Michlewski G, Guil S, Semple CA, Céceres JF . Posttranscriptional regulation of miRNAs harboring conserved terminal loops . _Mol Cell_ 2008 ; 32 :
383 - 393 Article CAS PubMed PubMed Central Google Scholar * Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ . Pathogenic roles for Epstein-Barr virus (EBV) gene products in
EBV-associated proliferative disorders . _Crit Rev Oncol Hematol_ 2003 ; 45 : 1 - 36 Article PubMed Google Scholar * Minarovits J, Minarovits-Kormuta S, Ehlin-Henriksson B, Falk K, Klein
G, Ernberg I . Host cell phenotype-dependent methylation patterns of Epstein-Barr virus DNA . _J Gen Virol_ 1991 ; 72 : 1591 - 1599 Article CAS PubMed Google Scholar * O'Donnell KA,
Wentzel EA, Zeller KI, Dang CV, Mendell JT . c-Myc-regulated microRNAs modulate E2F1 expression . _Nature_ 2005 ; 435 : 839 - 843 Article CAS PubMed Google Scholar * Oh ST, Cha JH, Shin
DJ, Yoon SK, Lee SK . Establishment and characterization of an _in vivo_ model for Epstein-Barr virus positive gastric carcinoma . _J Med Virol_ 2007 ; 79 : 1343 - 1348 Article CAS PubMed
Google Scholar * Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD . DNA methylation inhibitor 5-Aza-2'-deoxycytidine induces reversible genome-wide DNA damage that is
distinctly influenced by DNA methyltransferases 1 and 3B . _Mol Cell Biol_ 2008 ; 28 : 752 - 771 Article CAS PubMed Google Scholar * Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ,
Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T . Identification of virus-encoded microRNAs . _Science_ 2004 ; 304 : 734 - 736 Article CAS PubMed Google Scholar * Pratt ZL,
Kuzembayeva M, Sengupta S, Sugden B . The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines . _Virology_ 2009 ; 386 : 387 - 397 Article CAS
PubMed Google Scholar * Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF . Myogenic factors that regulate expression of muscle-specific microRNAs . _Proc Natl Acad Sci U S A_ 2006
; 103 : 8721 - 8726 Article CAS PubMed PubMed Central Google Scholar * Robertson KD, Hayward SD, Ling PD, Samid D, Ambinder RF . Transcriptional activation of the Epstein-Barr virus
latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial . _Mol Cell Biol_ 1995 ; 15 : 6150 - 6159 Article CAS PubMed PubMed Central
Google Scholar * Robertson KD, Ambinder RF . Mapping promoter regions that are hypersensitive to methylation-mediated inhibition of transcription: application of the methylation cassette
assay to the Epstein-Barr virus major latency promoter . _J Virol_ 1997 ; 71 : 6445 - 6454 CAS PubMed PubMed Central Google Scholar * Saito Y, Liang G, Egger G, Friedman JM, Chuang JC,
Coetzee GA, Jones PA . Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells . _Cancer Cell_ 2006 ; 9 : 435 -
443 Article CAS PubMed Google Scholar * Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G . Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene
EGFL7 in human cancer cells . _Biochem Biophys Res Commun_ 2009 ; 379 : 726 - 731 Article CAS PubMed Google Scholar * Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC . Rapid alteration
of microRNA levels by histone deacetylase inhibition . _Cancer Res_ 2006 ; 66 : 1277 - 1281 Article CAS PubMed Google Scholar * Seo JS, Cho NY, Kim HR, Tsurumi T, Jang YS, Lee WK, Lee SK
. Cell cycle arrest and lytic induction of EBV-transformed B lymphoblastoid cells by a histone deacetylase inhibitor, Trichostatin A . _Oncol Rep_ 2008 ; 19 : 93 - 98 CAS PubMed Google
Scholar * Smith P . Epstein-Barr virus complementary strand transcripts (CSTs/BARTs) and cancer . _Semin Cancer Biol_ 2001 ; 11 : 469 - 476 Article CAS PubMed Google Scholar * Smith PR,
Gao Y, Karran L, Jones MD, Snudden D, Griffin BE . Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors . _J Virol_ 1993 ; 67 : 3217 - 3225
CAS PubMed PubMed Central Google Scholar * Smith PR, de Jesus O, Turner D, Hollyoake M, Karstegl CE, Griffin BE, Karran L, Wang Y, Hayward SD, Farrell PJ . Structure and coding content
of CST (BART) family RNAs of Epstein-Barr virus . _J Virol_ 2000 ; 74 : 3082 - 3092 Article CAS PubMed PubMed Central Google Scholar * Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa
M, Okamoto K, Osato T . Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells . _Br J Cancer_ 1996 ; 74
: 625 - 631 Article CAS PubMed PubMed Central Google Scholar * Tao Q, Robertson KD . Stealth technology: how Epstein-Barr virus utilizes DNA methylation to cloak itself from immune
detection . _Clin Immunol_ 2003 ; 109 : 53 - 63 Article CAS PubMed Google Scholar * Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T . Epigenetic silencing of
microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer . _Cancer Res_ 2008 ; 68 : 4123 - 4132 Article CAS PubMed Google Scholar *
van Beek J, Brink AA, Vervoort MB, van Zijp MJ, Meijer CJ, van den Brule AJ, Middeldorp JM . _In vivo_ transcription of the Epstein-Barr virus (EBV) BamHI-A region without associated _in
vivo_ BARF0 protein expression in multiple EBV-associated disorders . _J Gen Virol_ 2003 ; 84 : 2647 - 2659 Article CAS PubMed Google Scholar * Viswanathan SR, Daley GQ, Gregory RI .
Selective blockade of microRNA processing by Lin28 . _Science_ 2008 ; 320 : 97 - 100 Article CAS PubMed PubMed Central Google Scholar * Wei MX, Moulin JC, Decaussin G, Berger F, Ooka T
. Expression and tumorigenicity of the Epstein-Barr virus BARF1 gene in human Louckes B-lymphocyte cell line . _Cancer Res_ 1994 ; 54 : 1843 - 1848 CAS PubMed Google Scholar * Winter J,
Jung S, Keller S, Gregory RI, Diederichs S . Many roads to maturity: microRNA biogenesis pathways and their regulation . _Nat Cell Biol_ 2009 ; 11 : 228 - 234 Article CAS PubMed Google
Scholar * Xing L, Kieff E . Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication . _J Virol_ 2007 ; 81 : 9967 - 9975 Article CAS PubMed
PubMed Central Google Scholar * Young LS, Rickinson AB . Epstein-Barr virus: 40 years on . _Nat Rev Cancer_ 2004 ; 4 : 757 - 768 Article CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS This work was supported by grants from the Gyeonggi Regional Research Centre (GRRC) of the Catholic University of Korea [(GRRC Catholic 2010 - A01). RNA-based development of
biopharmaceutical lead molecules] and from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family affairs, Republic of Korea (0920210). AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Department of Medical Lifescience, Research Institute of Immunobiology, College of Medicine, The Catholic University of Korea, Seoul 137-701, Korea., Do Nyun Kim
& Suk Kyeong Lee * Department of Life Science, Kyungwon University, Seongnam 461-701, Korea., Yoon-Jae Song Authors * Do Nyun Kim View author publications You can also search for this
author inPubMed Google Scholar * Yoon-Jae Song View author publications You can also search for this author inPubMed Google Scholar * Suk Kyeong Lee View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Suk Kyeong Lee. ADDITIONAL INFORMATION Supplementary Information accompanies the paper on the
Experimental & Molecular Medicine website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (PDF 211 KB) RIGHTS AND PERMISSIONS This is an Open Access article distributed under the
terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and
reproduction in any medium, provided the original work is properly cited. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kim, D., Song, YJ. & Lee, S. The role of promoter
methylation in Epstein-Barr virus (EBV) microRNA expression in EBV-infected B cell lines. _Exp Mol Med_ 43, 401–410 (2011). https://doi.org/10.3858/emm.2011.43.7.044 Download citation *
Accepted: 25 May 2011 * Published: 01 June 2011 * Issue Date: July 2011 * DOI: https://doi.org/10.3858/emm.2011.43.7.044 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * decitabine * DNA methylation * Herpesvirus 4, human * microRNAs * promoter regions, genetic
:max_bytes(150000):strip_icc():focal(216x0:218x2)/benedict-cumberbatch-1-435-4-20cc736017b24435a3498a49d7c22b0e.jpg)