- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT _MLL2_ mutations are detected in 55 to 80% of patients with Kabuki syndrome (KS). In 20 to 45% patients with KS, the genetic basis remains unknown, suggesting possible genetic
heterogeneity. Here, we present the largest yet reported cohort of 116 patients with KS. We identified _MLL2_ variants in 74 patients, of which 47 are novel and a majority are truncating. We
show that pathogenic missense mutations were commonly located in exon 48. We undertook a systematic facial KS morphology study of patients with KS at our regional dysmorphology meeting. Our
data suggest that nearly all patients with typical KS facial features have pathogenic _MLL2_ mutations, although KS can be phenotypically variable. Furthermore, we show that _MLL2_
mutation-positive KS patients are more likely to have feeding problems, kidney anomalies, early breast bud development, joint dislocations and palatal malformations in comparison with _MLL2_
mutation-negative patients. Our work expands the mutation spectrum of _MLL2_ that may help in better understanding of this molecule, which is important in gene expression, epigenetic
control of active chromatin states, embryonic development and cancer. Our analyses of the phenotype indicates that _MLL2_ mutation-positive and -negative patients differ systematically, and
genetic heterogeneity of KS is not as extensive as previously suggested. Moreover, phenotypic variability of KS suggests that _MLL2_ testing should be considered even in atypical patients.
SIMILAR CONTENT BEING VIEWED BY OTHERS CLINICAL AND MOLECULAR CHARACTERISTICS OF KOREAN PATIENTS WITH KABUKI SYNDROME Article 01 June 2024 DEFINING THE GENOTYPIC AND PHENOTYPIC SPECTRUM OF
X-LINKED _MSL3_-RELATED DISORDER Article Open access 11 November 2020 CLINICAL DELINEATION OF SETBP1 HAPLOINSUFFICIENCY DISORDER Article 19 April 2021 INTRODUCTION Kabuki syndrome (KS,
Kabuki make-up syndrome or Niikawa–Kuroki syndrome) is an autosomal dominant condition that arises _de novo_ in a majority of cases and is characterised by a recognisable facial phenotype of
interrupted high-arched eyebrows, long palpebral fissures, eversion of lateral part of lower eyelids, broad depressed nasal tip, large prominent earlobes and pillowed lower lip.1, 2 All
children with KS are globally delayed and have mild-to-moderate learning disability (for review of clinical features, see Adam and Hudgins3). They also have a high incidence of internal
malformations, involving, but not limited to, the heart, kidneys, gastrointestinal system, skeletal system and eyes. KS is commonly associated with severe feeding difficulties, hypotonia,
postnatal failure to thrive, obesity in later childhood and frequent infections. Rare problems include idiopathic thrombocytopenic purpura, seizures, autistic traits, delayed puberty and
tumours. Recently, whole exome sequencing led to the identification of mutations in _MLL2_ as the basis of KS in a majority of the patients.4 Still, the underlying cause cannot be identified
in 20 to 45% of patients with a presumed diagnosis of KS, suggesting possible genetic heterogeneity.5, 6, 7, 8 To investigate the spectrum of mutations associated with KS, we sequenced
_MLL2_ in 116 patients with clinically suspected KS. To test if KS is genetically heterogeneous, we systematically evaluated differences between the phenotype of patients in whom _MLL2_
mutations were found _versus_ those in whom mutations were not detected. PATIENTS AND METHODS PATIENT ASCERTAINMENT Before the discovery of _MLL2_ mutations as a cause of KS, we had
ascertained a highly selected group of ∼35 patients, based on clinical features and facial dysmorphology, through the Clinical Genetics clinics in Manchester. Of these 35 patients, 18 were
included in the study reported by Hannibal _et al._8 After the gene discovery, we expanded our cohort by accepting patients with possible KS from Clinical Genetics departments, mostly across
the United Kingdom. Referring clinicians were asked to provide DNA samples from their patients and, if possible/required, from both parents. A clinical proforma was designed to gather
phenotypic information from the referring clinicians. Photographs of patients were also collected wherever possible. The study was approved by the Central Manchester Research Ethics
Committee (02/CM/238) and the University of Manchester Ethics Committee. Fully informed signed consents were taken from legal guardians of the patients by the referring clinicians. The study
was performed in accordance with the Declaration of Helsinki protocol. MUTATION SCREENING OF _MLL2_ GENE All the patients were screened for mutations in the _MLL2_ gene. PCR amplification
for all the 54 exons spanning the _MLL2_ gene was performed using primer sequences and the PCR conditions previously described.4 PCR amplification was carried out on Veriti thermal cycler or
2720 thermal cycler (Applied Biosystems, Paisley, UK) using the Reddy Mix Custom PCR Master Mix (ABgene, Epsom, UK, catalogue no: AB-0575/DC/LD/b) according to the manufacturer's
instructions. Amplified products were cleaned using Agencourt AMPure XP (Beckman Coulter Genomics, Takeley, UK) system on an automated Beckman Coulter Liquid Handler, Biomek 3000, as per the
manufacturer's instructions. The purified PCR products were then subjected to direct sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). All the
sequencing reactions were cleaned using Agencourt CleanSeq on automated Beckman Coulter Liquid Handler and were then sequenced in the departmental core facility on an ABI prism 3100 Genetic
Analyser (Applied Biosystems). The sequence data generated were exported and analysed graphically using the DNA sequence analysis software, STADEN (http://staden.sourceforge.net/). The
changes observed were checked against the NCBI SNP database to identify known single-nucleotide polymorphisms (SNPs). Sequence variations found were reported according to the Human Genome
Variation Society nomenclature. Where available, DNA from the parents of the patients with mutations was screened to investigate if the change was _de novo_ or inherited. All the mutations
that were identified in this study and in our patients reported by Hannibal _et al_8 were independently confirmed. We designed new primer sets for this step to avoid amplicons of >700
base pairs. These primer sequences are available on request. FACIAL KS MORPHOLOGY SCORE Clinical geneticists and dysmorphologists from Manchester, Liverpool, Leeds, Newcastle upon Tyne,
Sheffield and Cardiff meet six monthly in Manchester at ‘M62 dysmorphology meeting’ to discuss unusual cases. We decided to test if facial dysmorphism can be a reliable predictor of _MLL2_
mutation status in our cohort. We showed 37 facial photographs of 33 patients from our cohort to 23 dysmorphologists who had gathered for the M62 meeting in June 2011. Photographs for 15
_MLL2-_negative patients were available. Photographs of all these 15 patients were included in the study to eliminate selection bias and provide enough statistical power. Although more
photographs of patients with definite _MLL2_ mutation were available, only 18 were included in the study. This was done to ensure that the presentation lasted for <10 min to avoid
participant boredom that could adversely affect the results. Majority of the photographs for _MLL2_ mutation-positive patients were selected randomly, but some with atypical appearance were
included intentionally to test the whole spectrum of facial dysmorphism of KS. Participants were blind to clinical histories and mutation status of the patients. Each photograph was shown
for 15 s and participants were asked to rate the likelihood of the patient having KS on an ordinal scale of one to five (one for ‘unlikely KS’, three for ‘possible KS’ and five for ‘definite
KS’). The results were analysed using Mann–Whitney test. To assess if facial KS morphology scores were age dependent, we included two photographs taken at different ages for four patients.
GENOTYPE–PHENOTYPE ANALYSIS Detailed information on clinical features was available for 63/116 patients (40 _MLL2_ mutation positive and 23 _MLL2_ mutation negative). We concentrated on
analysing differences between those features where information was reliably quantifiable and was available in majority of the cases. Therefore, we did not analyse differences between growth,
developmental delay and learning disability. Two-tailed _P-_values were generated by Fisher's exact test. RESULTS MUTATION ANALYSIS We sequenced all 54 exons and the exon–intron
boundaries in 98 patients with a suspected diagnosis of KS. For the purpose of analysis, we have also included results from our additional 18 patients (giving a total of 116) who have
already been reported by Hannibal _et al._8 All the patients previously reported in another study have been identified clearly in the comments box of each table (Supplementary Tables 1–4).
We ascribed ‘definite pathogenicity’ only to mutations resulting in truncated protein or obvious splicing error or missense changes that were proven to be _de novo_ in at least one patient.
If both parental samples were unavailable, a missense change was considered to be ‘likely pathogenic’ if it has been identified in more than one patient and _in silico_ predictions indicated
high degree of conservation and disruption of protein function. Changes that were inherited from an unaffected parent or found concurrently in the same patient with another definitely
pathogenic variant were assigned as ‘unlikely pathogenic’. Changes that could not be qualified to any of these categories were labelled to be of ‘uncertain pathogenicity’. Following our
criteria, no pathogenic variants were found in 42/116 (36.2%) patients. In 74/116 (63.8%) patients, we identified _MLL2_ variants, out of which 66 variants were inferred to be definite
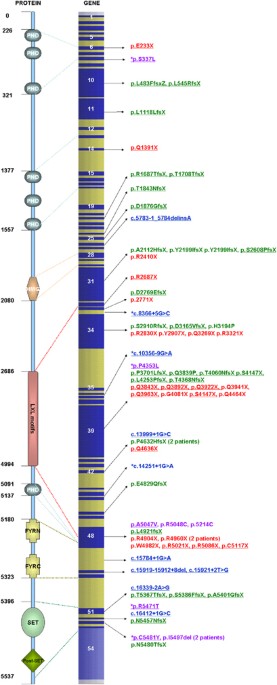
disease causing and 2 to be likely pathogenic (Figure 1). We could not be certain of the significance of six changes. Out of 74 changes, 25 were nonsense mutations, 21 were frameshift
deletions, 9 were frameshift insertions, 10 were splice-site variants, 9 were missense changes and 2 were in-frame deletions. Both the changes classified as likely pathogenic were missense
changes. Out of six changes with uncertain significance, two potentially affected splicing and four were missense changes. In addition, 47 changes were novel variants. FACIAL KS MORPHOLOGY
SCORES Mean and median values for facial KS morphology scores were calculated for each patient. We considered an average score of >3.5 and <3 as being indicative of features highly
compatible or incompatible with KS, respectively. For scores between 3 and 3.5, we assumed that facial dysmorphism was not enough to strongly suggest or reject KS as a diagnosis for the
individual. Table 1 provides a summary of facial KS morphology scores in all 33 patients. Our analysis showed that 13/15 _MLL2_ mutation-negative patients had a mean score of <3 for
facial KS morphology, whereas 12/18 _MLL2_ mutation-positive patients scored >3.5. Although there is some overlap between the scores of two groups, the difference between facial KS
morphology scores of _MLL2_ mutation-positive and -negative KS patients was highly significant, with the _P-_value of <0.0001. GENOTYPE–PHENOTYPE CORRELATION We analysed our phenotype
data to investigate if there were any differences between other clinical features of _MLL2_ mutation-positive and -negative groups. The results showed that feeding problems, urogenital
anomalies, early breast buds, joint dislocations and cleft palate were significantly more common in patients with _MLL2_ mutations (Table 2). DISCUSSION MLL2 is a histone H3 lysine 4
(H3K4)-specific methyl transferase that belongs to the SET1 family of human SET-domain protein methyltransferase superfamily.9, 10 It is a large protein that consists of seven plant
homeodomains (PHD, for protein–protein interaction), one high-mobility group domain (for binding to DNA with low sequence specificity), five LXXLL motifs (for interaction with nuclear
receptors), one each FYRC and FYRN domains (for heterodimerisation between terminal fragments of MLL) and a single SET domain (for histone lysine methylation) (Figure 1).10 MLL2 functions as
a part of multiprotein complex, ASCOM, which binds to the regulatory sites of the target genes.11 It is important for epigenetic transcriptional activation, it interacts with oestrogen
receptor-_α_ and is important for embryonic development.9, 12 We identified variants in 74/116 (63.8%) patients with KS in our study (Figure 1). Ng _et al_4 discovered mutations in 35/53
(66%) cases of KS through a combination of exome and Sanger sequencing. Paulussen _et al_,5 Li _et al_6 and Micale _et al_7 identified _MLL2_ mutations in 34/45 (75.5%), 19/34 (55.8%) and
45/62 (72.5%) patients, respectively. Most recently, Hannibal _et al_8 reported 57 more patients with KS and found mutations in 46 individuals (80%), including 17 of our 18 patients. We
present here the largest cohort of _MLL2_ mutation-positive patients with KS identified as yet. Overall mutation detection rate between different studies is variable, which is likely a
reflection of differences in ascertainment. Including this study, there are now 232 KS kindreds with _MLL2_ mutations in the published literature. We have analysed the _MLL2_ mutation
spectrum by combining our data with what is already published. TRUNCATING MUTATIONS Out of 232 published _MLL2_ mutation-positive KS patients, including 55 patients reported here, 170
(73.2%) have truncating mutations (Supplementary Table 1). A number of these mutations have now been reported in more than one patient and are highlighted in the Supplementary Table 1. Out
of 170, 86 (more than half) are nonsense mutations. In addition, 59 patients have deletions, with the majority of a single base. The biggest reported deletion is of 32 bases. Notably,
c.6595delT (p.Y2199IfsX65) in exon 31 has been described in five patients, including two patients in our study. It has been shown to be _de novo_ in three patients, confirming that it is a
recurrent mutation. Also, 22 patients have small duplications or insertions. Three patients have complicated insertion and duplication, with two involving exon 34. Of note, a number of exons
are disproportionately affected by truncating mutations. For example, exon 52, which codes for 0.44% of the translated transcript, accounts for three mutations in four patients, more than
five times the expected rate. Exons 27, 33, 40 and 53 also account for excessively large number of truncating mutations, although the sample size is insufficient to draw statistical
conclusions. The largest numbers of truncating mutations have been found in exon 39 (36 mutations in 39 patients) and exon 48 (16 mutations in 21 patients). Exons 39 and 48 code for 16.8 and
6.9% of MLL2 protein, correspondingly. The truncating mutation closest to N and C terminals of the protein are p.C158VfsX50 (exon 4) and p.R5501X (exon 53). SPLICE-SITE MUTATIONS We have
added 10 splice-site mutations (9 novel) to the list of _MLL2_ mutations. Including patients described in our study, 21 splice-site mutations are now described in patients with KS
(Supplementary Table 2). One patient in our study has a deletion spanning from the end of exon 49 to 10 bases into the intron, resulting in deletion of a single amino acid and abolishment of
a splice donor site. Out of 19 _MLL2_ splice-site mutations, 7 have been shown to be _de novo_. Out of 10 splice variants, 2 are of uncertain pathogenicity. Resultant proteins of none of
the _MLL2_ splice-site mutations have been characterised as yet. There are no obvious phenotypic differences between patients with splice-site and other mutations. Notably, one patient with
c.13999+1G developed low-grade myofibrosarcoma and has been reported earlier.13 NONTRUNCATING MUTATIONS A total of 28 missense or in-frame insertions or deletions in _MLL2_, including 9 in
this study, have been reported in 37 patients with KS. In our study, only those nontruncating changes that were proven to be _de novo_ in at least one patient were considered to be
definitely pathogenic. Using the same criteria in all the reported mutations, we can be certain about deleterious effect of 13 mutations in 22 patients (Supplementary Table 3). In 15
patients, 15 changes have not been checked for inheritance. It is therefore not possible to be certain about their pathogenicity. Out of these 28 changes, one is a duplication of two amino
acids resulting from a six base pair duplication in exon 48.7 p.I5497del, a single amino-acid deletion within the SET domain resulting from a mutation in exon 53, has been described in three
patients, including one in this study. In our patient, this change was shown to be _de novo_ and hence certainly pathogenic. Other 26/28 changes are missense. Most of the pathogenic
missense changes are concentrated toward the −COOH terminal of the protein. Out of the 37 (43.2%) patients with non-truncating non-splice-site mutations, 16 have changes in exon 48 (that
codes for <7% of MLL2 protein). Five missense mutations in exon 48 have been described in more than one patient. This includes p.R5048C mutation that we found in a previously reported
mother–daughter pair.14 Of note, exon 48 codes for LXXL motifs, one PHD domain and part of the FYRN domain. We have also compiled a list of changes that are not known polymorphisms but have
been seen concurrently in patients who have another definite pathogenic mutation (Supplementary Table 4). These are unlikely to be pathogenic but may be useful in understanding the role of
various MLL2 residues. In this list we have also included variants that have been shown to be inherited from a normal parent. In our opinion, in the absence of stronger evidence to suggest
variable penetrance in KS, such variants should be provisionally considered as benign. This degree of caution is especially important in clinical practice. Of note, four out of the five
variants that we found concurrently with another definitely pathogenic mutation in this study are predicted to be highly damaging by AlignGVD and SIFT. This shows that _in silico_
predictions for MLL2 are currently not entirely reliable, which is probably because the structure of MLL2 has not yet been realised. Hence, in a patient with KS, it is important to establish
that a missense _MLL2_ change has arisen _de novo_ before ascribing definite pathogenicity. FACIAL KS MORPHOLOGY SCORES Our group had contributed 18 patients to the study reported by
Hannibal _et al_8 and, interestingly, _MLL2_ mutations were found in 17/18 patients (94.4%). Notably, these 18 patients were selected for having clinical features that are considered to be
most typical of KS. The characteristics of mutations found in this group did not differ in any way from the larger cohort. Most previous studies have proposed that KS is genetically
heterogeneous, but our experience indicates that most typical cases of KS are indeed due to _MLL2_ mutations. Furthermore, no strong genotype–phenotype correlation has emerged in previous
studies comparing _MLL2_ mutation-positive and -negative patients. We hypothesised that facial KS morphology might be the most important difference between the two groups. We undertook a
systematic approach to score facial KS morphology to test the genetic heterogeneity of ‘true’ KS. Results of our facial KS morphology scores show that nearly all patients with ‘typical
Kabuki face’ have mutations in _MLL2_ (Figures 2a, b and Table 1). Furthermore, there was no apparent trend correlating facial KS morphology scores with the type of mutation seen. Only 1 out
of 15 _MLL2_ mutation-negative KS patients (KS110) scored high on the facial KS morphology score (93/115, mean 4.04; Figure 2d). Clinical history of this patient was also reminiscent of
typical KS. Further work is needed to identify the underlying molecular mechanism in this child. Overall, this shows that based on facial dysmorphism, genetic heterogeneity of KS is not
likely to be as large as previously expected. A few patients who had relatively low facial KS morphology scores are not absolutely facially typical for KS but still have _MLL2_ mutations,
demonstrating variability of facial phenotype in KS (see KS 62 and 36 in Table 1; Figure 2c). This indicates that _MLL2_ testing should be considered even in atypical KS patients. In our and
other geneticists' experience, it can be difficult to recognise KS in early infancy and in adulthood. Interestingly, for four patients whose two photographs taken at different ages
were included in the slide show (Figures 3a–d), the scores for early childhood photographs corresponded better to the mutation status than those taken in infancy or adulthood. Overall, the
relationship between facial dysmorphism and _MLL2_ mutation status seems compelling, but these results should be interpreted with caution because by nature, such studies are subjective and
open to many biases. GENOTYPE–PHENOTYPE CORRELATION Our finding of significant differences between feeding difficulties, early breast bud development or gynaecomastia, joint dislocations and
cleft palate adds further to help delineate features that may increase the chances of finding _MLL2_ mutation in a patient (Table 2). Similar to two previous studies,6, 8 we also observed
significant differences in rates of urogenital malformations. The range of feeding difficulties in MLL2 mutation-positive patients varied from just slow feeding or gastro-oesophageal reflux
to needing nasogastric tube feeding and/or even gastrostomy. Most frequent joint dislocations were reported to involve the patella, hips, shoulders and elbows. One patient had bilateral
thumb subluxations. A variety of palatal malformations like bifid or absent uvula, velopharyngeal insufficiency and central or bilateral cleft palate were seen in _MLL2_ mutation-positive
patients. _MLL2_ MUTATION-NEGATIVE PATIENTS Deep intronic mutations, large intragenic deletions or duplications or changes affecting the regulatory elements may not be recognised by
sequencing of exons and exon–intron boundaries. Perhaps, MLPA or mRNA sequencing should be considered in typical _MLL2_ mutation-negative KS patients. However, from the facial KS morphology
scores and emerging genotype phenotype correlation, it seems likely that many suspected KS patients without _MLL2_ mutation are likely to have unrelated conditions that may not be due to
defects even in the same pathway. This is further indicated by the failure to find mutations in any genes associated with ASCOM complex in _MLL2_ mutation-negative patients in two other
studies.6, 8 Four _MLL2_ mutation-negative patients with proposed KS in the study reported by Hannibal _et al_8 were found to have an alternative diagnosis by array comparative genomic
hybridisation (a-CGH). One patient had a deletion on 5q encompassing _NSD1_, the second had a novel 19q23 deletion involving 20 genes, the third had a complex translocation involving
chromosomes 8 and 18 and the fourth patient had mosaic trisomy 12. In our cohort we have not systematically performed a-CGH on all _MLL2_ mutation-negative patients but have found a deletion
of 6q13, inherited from her affected mother, in one patient who we have always regarded to have atypical KS. Her mother also has similar clinical features. Similar deletions have been
described in the literature.15 We therefore suggest a-CGH should be considered in _MLL2_ mutation-negative KS patients, if not done already. CONCLUSIONS Our work expands the known mutation
spectrum of _MLL2_ mutations associated with KS. We have shown that the majority of mutations are truncating and the pathogenic missense mutations are commonly located in exon 48. We suggest
that caution should be practised in assigning pathogenicity to missense _MLL2_ variants. Our compilation of mutation data available from the published literature may help laboratories
offering _MLL2_ testing to ascribe significance to novel findings. Our mutation detection rate of ∼60% is a closer reflection of expected rate in clinical setting. Overall, this may provide
insight into the structure and function of MLL2. We show that KS can be phenotypically variable and therefore _MLL2_ testing should be considered even in atypical KS patients. Furthermore,
we have demonstrated that _MLL2_ mutation-positive and -negative patients differ significantly in their facial dysmorphism and some clinical features. The majority of typical KS patients
have _MLL2_ mutations, implying that the genetic heterogeneity of KS may be minimal. This work will also help in further studies to identify the genetic basis of patients with MLL2
mutation-negative KS. REFERENCES * Kuroki Y, Suzuki Y, Chyo H, Hata A, Matsui I : A new malformation syndrome of long palpebralfissures, large ears, depressed nasal tip, and skeletal
anomalies associated with postnatal dwarfism and mental retardation. _J Pediatr_ 1981; 99: 570–573. Article CAS Google Scholar * Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T :
Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. _J Pediatr_ 1981; 99: 565–569. Article CAS Google
Scholar * Adam MP, Hudgins L : Kabuki syndrome: a review. _Clin Genet_ 2005; 67: 209–219. Article CAS Google Scholar * Ng SB, Buckingham KJ, Hannibal MC _et al_: Exome sequencing
identifies MLL2 mutations as a cause of Kabuki syndrome. _Nat Genet_ 2010; 42: 790–793. Article CAS Google Scholar * Paulussen ADC, Stegmann APA, Blok MJ _et al_: MLL2 mutation spectrum
in 45 patients with Kabuki syndrome. _Hum Mutat_ 2011; 32: E2018–E2025. Article CAS Google Scholar * Li Y, Bögershausen N, Alanay Y _et al_: A mutation screen in patients with Kabuki
syndrome. _Hum Genet_ 2011; 130: 715–724. Article CAS Google Scholar * Micale L, Augello B, Fusco C _et al_: Mutation spectrum of MLL2 in a cohort of Kabuki syndrome patients. _Orphanet J
Rare Dis_ 2011; 6: 38. Article Google Scholar * Hannibal MC, Buckingham KJ, Ng SB _et al_: Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. _Am J Med Genet_ 2011; 155A:
1511–1516. Article Google Scholar * Dillon S, Zhang X, Trievel R, Cheng X : The SET-domain protein superfamily: protein lysine methyltransferases. _Genome Biol_ 2005; 6: 227. Article
Google Scholar * Ansari KI, Mandal SS : Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. _FEBS J_ 2010; 277: 1790–1804. Article CAS Google Scholar
* Goo Y-H, Sohn YC, Kim D-H _et al_: Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. _Mol Cell Biol_ 2003; 23:
140–149. Article CAS Google Scholar * Mo R, Rao SM, Zhu Y-J : Identification of the MLL2 complex as a coactivator for estrogen receptor α. _J Biol Chem_ 2006; 281: 15714–15720. Article
CAS Google Scholar * Shahdadpuri R, O'Meara A, O'Sullivan M, Reardon W : Low-grade fibromyxoid sarcoma: yet another malignancy associated with Kabuki syndrome. _Clin Dysmorphol_
2008; 17: 199–202. Article Google Scholar * Pottinger CC, Stiff RE, Holroyd JM, Davies SJ : Further evidence of dominant inheritance of Kabuki syndrome. _Clin Dysmorphol_ 2009; 18:
215–217. Article Google Scholar * Van Esch H, Rosser EM, Janssens S _et al_: Developmental delay and connective tissue disorder in four patients sharing a common microdeletion at 6q13-14.
_J Med Genet_ 2010; 47: 717–720. Article Google Scholar Download references ACKNOWLEDGEMENTS We are grateful to Mr and Mrs P Scales, their family and friends for funding and support
received for this project through the Central Manchester University Hospitals NHS Foundation Trust, Kabuki Research Fund no. 629396. We also acknowledge the support of Manchester Biomedical
Research Centre. We would like to show our appreciation to Dr Michael J Bamshad and Dr Katie J Buckingham from Seattle for providing us information for the primers for _MLL2_ sequencing. We
thank all our patients, families and their clinicians for help in the research. We are grateful to following participants for scoring the facial KS morphology of patients at the M62
dysmorphology meeting: Dr Miranda Splitt, Dr Brian Wilson and Dr Gaik Siew (Newcastle upon Tyne); Dr Alan Fryer, Dr Lynn Greenhalgh and Dr Elizabeth Sweeney (Liverpool); Dr Angus Dobie, Dr
Claire Searle, Dr Alison Krauss and Dr Jenny Thomson (Leeds); Dr Diana Johnson and Dr Meena Balasubramanian (Sheffield); and Dr Elizabeth Jones and Dr Sofia Douzgou (Manchester). AUTHOR
INFORMATION Author notes * Siddharth Banka and Ratna Veeramachaneni: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Genetic Medicine, St Mary's
Hospital, Manchester Academic Health Sciences Centre (MAHSC), University of Manchester, Manchester, UK Siddharth Banka, Ratna Veeramachaneni, Emma Howard, Sancha Bunstone, Yanick J Crow,
Bronwyn Kerr, Helen Kingston, Kay Metcalfe, Kate Chandler, Jill Clayton-Smith, Graeme C Black & Dian Donnai * National Centre for Medical Genetics, Our Lady's Children's
Hospital, Dublin, Ireland William Reardon, Sally Ann Lynch & Harinder Gill * Wessex clinical Genetics Service, University Hospital Southampton NHS Trust and Human Genetics and Genomic
Medicine, Faculty of Medicine, University of Southampton, Southampton, UK Nicola Ragge, I Karen Temple, Amanda L Collins & Katherine Lachlan * Department of Clinical Genetics, Sheffield
Children's NHS Foundation Trust, Sheffield, UK Michael J Parker * Northern Ireland Regional Genetics Service, Belfast City Hospital, Belfast, UK Alex Magee, Fiona Stewart, Vivienne P M
McConnell & Deirdre E Donnelly * Center for Medical Genetics and Molecular Medicine, Haukeland University Hospital, Bergen, Norway Siren Berland & Gunnar Houge * Clinical Genetics
Unit, Birmingham Women's Hospital, Birmingham, UK Jenny E Morton & Christine Oley * Center for Human Genetics, Cliniques universitaires Saint-Luc, Université catholique de Louvain,
Brussels, Belgium Nicole Revencu * Department of Clinical Genetics, East Anglian Medical Genetics Service, Addenbrooke's Hospital, Cambridge, UK Soo-Mi Park * All Wales Medical Genetics
Service, Institute of Medical Genetics, University Hospital of Wales, Cardiff, UK Sally J Davies & Andrew E Fry * Division of Medical Sciences, University of Dundee, Dundee, UK Susann
Schweiger * South East of Scotland Clinical Genetic Service, Molecular Medicine Centre, Western General Hospital, Edinburgh, UK Wayne W K Lam * Clinical Genetics Department, Yorkhill
Hospitals, Glasgow, UK John Tolmie * Clinical Genetics, Guy's Hospital, London, UK Shehla N Mohammed * Yorkshire Regional Genetic Service, Leeds, UK Emma Hobson, Audrey Smith, Moira
Blyth & Christopher Bennett * Department of Clinical Genetics, Leicester Royal Infirmary Leicester, UK Pradeep C Vasudevan * Instituto de Genética Médica y Molecular (INGEMM), Hospital
Universitario La Paz, Madrid, Spain Sixto García-Miñaúr * Northern Genetics Service, Newcastle upon Tyne Hospitals, Newcastle upon Tyne, UK Alex Henderson, Judith Goodship & Michael J
Wright * Northern Genetics Service, Teesside Genetics Unit, The James Cook University Hospital, Middlesbrough, UK Richard Fisher * Department of Clinical Genetics, Churchill Hospital,
Oxford, UK Richard Gibbons & Susan M Price * Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka Deepthi C de Silva * Institute of Genetic Medicine, Newcastle University,
Newcastle upon Tyne, UK Judith Goodship * Department of Medical Genetics, St George's Hospital, London, UK Frances Elmslie & Meriel McEntagart * Clinical Genetics Department, Royal
Devon and Exeter Hospital, Exeter, UK Bruce Castle Authors * Siddharth Banka View author publications You can also search for this author inPubMed Google Scholar * Ratna Veeramachaneni View
author publications You can also search for this author inPubMed Google Scholar * William Reardon View author publications You can also search for this author inPubMed Google Scholar * Emma
Howard View author publications You can also search for this author inPubMed Google Scholar * Sancha Bunstone View author publications You can also search for this author inPubMed Google
Scholar * Nicola Ragge View author publications You can also search for this author inPubMed Google Scholar * Michael J Parker View author publications You can also search for this author
inPubMed Google Scholar * Yanick J Crow View author publications You can also search for this author inPubMed Google Scholar * Bronwyn Kerr View author publications You can also search for
this author inPubMed Google Scholar * Helen Kingston View author publications You can also search for this author inPubMed Google Scholar * Kay Metcalfe View author publications You can also
search for this author inPubMed Google Scholar * Kate Chandler View author publications You can also search for this author inPubMed Google Scholar * Alex Magee View author publications You
can also search for this author inPubMed Google Scholar * Fiona Stewart View author publications You can also search for this author inPubMed Google Scholar * Vivienne P M McConnell View
author publications You can also search for this author inPubMed Google Scholar * Deirdre E Donnelly View author publications You can also search for this author inPubMed Google Scholar *
Siren Berland View author publications You can also search for this author inPubMed Google Scholar * Gunnar Houge View author publications You can also search for this author inPubMed Google
Scholar * Jenny E Morton View author publications You can also search for this author inPubMed Google Scholar * Christine Oley View author publications You can also search for this author
inPubMed Google Scholar * Nicole Revencu View author publications You can also search for this author inPubMed Google Scholar * Soo-Mi Park View author publications You can also search for
this author inPubMed Google Scholar * Sally J Davies View author publications You can also search for this author inPubMed Google Scholar * Andrew E Fry View author publications You can also
search for this author inPubMed Google Scholar * Sally Ann Lynch View author publications You can also search for this author inPubMed Google Scholar * Harinder Gill View author
publications You can also search for this author inPubMed Google Scholar * Susann Schweiger View author publications You can also search for this author inPubMed Google Scholar * Wayne W K
Lam View author publications You can also search for this author inPubMed Google Scholar * John Tolmie View author publications You can also search for this author inPubMed Google Scholar *
Shehla N Mohammed View author publications You can also search for this author inPubMed Google Scholar * Emma Hobson View author publications You can also search for this author inPubMed
Google Scholar * Audrey Smith View author publications You can also search for this author inPubMed Google Scholar * Moira Blyth View author publications You can also search for this author
inPubMed Google Scholar * Christopher Bennett View author publications You can also search for this author inPubMed Google Scholar * Pradeep C Vasudevan View author publications You can also
search for this author inPubMed Google Scholar * Sixto García-Miñaúr View author publications You can also search for this author inPubMed Google Scholar * Alex Henderson View author
publications You can also search for this author inPubMed Google Scholar * Judith Goodship View author publications You can also search for this author inPubMed Google Scholar * Michael J
Wright View author publications You can also search for this author inPubMed Google Scholar * Richard Fisher View author publications You can also search for this author inPubMed Google
Scholar * Richard Gibbons View author publications You can also search for this author inPubMed Google Scholar * Susan M Price View author publications You can also search for this author
inPubMed Google Scholar * Deepthi C de Silva View author publications You can also search for this author inPubMed Google Scholar * I Karen Temple View author publications You can also
search for this author inPubMed Google Scholar * Amanda L Collins View author publications You can also search for this author inPubMed Google Scholar * Katherine Lachlan View author
publications You can also search for this author inPubMed Google Scholar * Frances Elmslie View author publications You can also search for this author inPubMed Google Scholar * Meriel
McEntagart View author publications You can also search for this author inPubMed Google Scholar * Bruce Castle View author publications You can also search for this author inPubMed Google
Scholar * Jill Clayton-Smith View author publications You can also search for this author inPubMed Google Scholar * Graeme C Black View author publications You can also search for this
author inPubMed Google Scholar * Dian Donnai View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Siddharth Banka.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies the paper on European Journal of Human
Genetics website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 410 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Banka, S.,
Veeramachaneni, R., Reardon, W. _et al._ How genetically heterogeneous is Kabuki syndrome?: _MLL2_ testing in 116 patients, review and analyses of mutation and phenotypic spectrum. _Eur J
Hum Genet_ 20, 381–388 (2012). https://doi.org/10.1038/ejhg.2011.220 Download citation * Received: 18 August 2011 * Revised: 13 October 2011 * Accepted: 20 October 2011 * Published: 30
November 2011 * Issue Date: April 2012 * DOI: https://doi.org/10.1038/ejhg.2011.220 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * Kabuki
syndrome * _MLL2_ * genetic heterogeneity * mutation spectrum * facial dysmorphism







:max_bytes(150000):strip_icc():focal(779x429:781x431)/Joan-Hotchkis-100422-2-e82ee6ab5c964fa59887c584378a1f0a.jpg)
