- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
DEAR EDITOR, Onconase, the smallest member of RNase A super family that was isolated from oocytes or early embryos of the Northern Leopard Frog (_Rana pipiens_), has been granted as an
orphan drug for the treatment of malignant mesothelioma by US FDA1,2. It was also tested in clinical trials for patients with non-small-cell lung cancer, breast cancer, and renal cell
cancer2. Onconase is extremely stable with a Tm of ∼87 °C, resists degradation by various proteases, and evades ribonulease inhibitors (RI) present in mammalian cell cytosol3,4,5, which
confers advantages to its application in cancer treatment. More importantly, onconase specifically induces apoptosis of cancer cells but has low cytotoxicity to their normal counterparts2.
One rational hypothesis for this selectivity is that onconase, a highly cationic protein with calculated pI > 9.5, is selectively internalized by tumor cells given that tumor cells
generally are more negatively charged than cognate normal cells2. Nevertheless, as an anti-cancer drug, the mechanisms of its antitumor activity are not well understood. The current model of
onconase-mediated cytotoxicity favors that onconase degrades tRNAs in tumor cells after its cytosolic internalization, which leads to ubiquitous inhibition of protein synthesis and
apoptosis2. However, increasing evidence indicates that degradation of tRNAs and inhibition of protein synthesis are not the sole cause of onconase-induced apoptosis6,7,8. Consistent with
the well-documented causal roles of microRNAs (miRNAs) in cancers, a recent study indicates that onconase regulates the expression of miRNAs in malignant pleural mesothelioma cell lines
(H2959, H2373, and H2591), and reveals that onconase controls cell proliferation, invasion, migration, and apoptosis through modulating miRNAs9. However, how onconase affects miRNA
expression remains unclear. Here our biochemical studies showed that miRNAs are the direct downstream RNA species of onconase. Intriguingly, we found that onconase downregulates miRNAs by
cleavage of miRNA precursors, thus reducing the amount of mature miRNAs produced from Dicer processing. Using recombinant onconase that was prepared as described previously10, we found that
miR-155 and miR-21, two well-known oncogenic miRNAs with high endogenous levels in mesothelioma cell line Msto-211h, were dose- and time-dependently downregulated by onconase in these cells
(Supplementary information, Figure S1A, S1B, S1D and S1E), accompanied by a significant upregulation of their respective targets examined, including miR-155 targets SOCS111, C/EBPβ12,13, and
ETS-114, and miR-21 targets PTEN15 and PDCD416 (Supplementary information, Figure S1C and S1F). Furthermore, our miRNA microarray assays showed that the majority of miRNAs in Msto-211h
cells were significantly downregulated by onconase (Supplementary information, Figure S2A), while our quantitative real-time PCR (qRT-PCR) assays validated the onconase-mediated
downregulation of let-7 family miRNAs, miR-21, miR-29a, miR-92a, miR-92b, miR-155, miR-221, and miR-222 (Supplementary information, Figure S2B). These results indicate that miRNA expression
is susceptible to onconase, consistent with the recent report9. However, different from the findings of upregulation of 5 miRNAs in addition to downregulation of the majority in mesothelioma
cells by onconase in that study9, we found that onconase appears to globally downregulate miRNAs in Msto-211h cells. This dissimilarity is likely due to a different cell line examined in
this study. Nevertheless, our microarray assays also showed that four miRNAs (miR-181a, miR-30a, miR-1280, and miR-720) were upregulated by onconase treatment (Supplementary information,
Figure S2A); however, further analyses revealed that all these four miRNAs showed low intensities in microarray assays (Supplementary information, Figure S2C). Moreover, our qRT-PCR assays
showed that miR-181a expression was actually reduced > 2-fold by onconase treatment (Supplementary information, Figure S2B), while the other three miRNAs were undetectable in qRT-PCR
assays (data not shown), implicating low endogenous levels of these miRNAs in Msto-211h cells and false positive in microarray assays. It is noteworthy that 4 out of 5 upregulated miRNAs
have extremely low values of intensities in their miRNA microarray data9, while all of these five upregulated miRNAs do not meet the cutoff of mean intensities above 500 in our microarray
assays. We next asked how onconase downregulates miRNAs. To this end, we performed _in vitro_ onconase reactions to examine the effect of onconase on chemically synthesized 23-nt mature
miR-155 and 65-nt pre-miR-155 strands (Supplementary information, Data S1). A human tRNAArg, which was transcribed by T7 RNA polymerase _in vitro_, was used as a positive control for
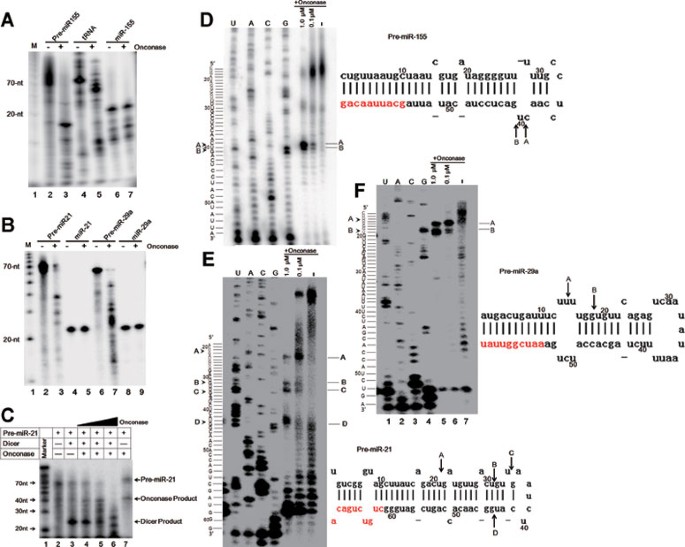
onconase activity. We found that this tRNA strand was strongly degraded by onconase (Figure 1A, lane 5), indicating that onconase prepared in the lab is highly active. Interestingly, we
found that miR-155 precursor strand was significantly cleaved by onconase (Figure 1A, lane 3), while mature miR-155 strand was marginally affected (Figure 1A, lane 7), suggesting that
onconase preferentially degrades the precursor forms of miRNAs. To further corroborate this, we examined the effect of onconase on other miRNAs. Consistently, we found that onconase strongly
degraded the precursor strands of miR-21 and miR-29a, but mildly affected their mature forms (Figure 1B). These results together indicate that oncoanse preferentially degrades miRNA
precursors instead of the mature forms of miRNAs. Given that onconase preferably cleaves miRNA precursors (Figure 1A and 1B) and that miRNA precursors are processed into miRNAs by Dicer in
cells, we speculated that onconase might reduce miRNA expression through disturbing Dicer-mediated mature miRNA production in cancer cells. Indeed, using _in vitro_ Dicer processing analyses
(Supplementary information, Data S1), we found that processing of pre-miR-21 into mature miR-21 by Dicer was dose-dependently reduced by onconase (Figure 1C). These results suggest that
onconase degrades miRNA precursors and subsequently results in less production of mature miRNAs produced by Dicer. Finally, we mapped the cleavage sites of onconase in miRNA precursors.
Using primer extension analyses (Supplementary information, Data S1), we found that pre-miR-155 was mainly cleaved at C39-U40 and U40-G41 (Figure 1D), pre-miR-21 at U21-G22, U31-G32,
U34-G35, and U43-G44 (Figure 1E), and pre-miR-29a at U14-U15 and U18-G19 (Figure 1F). These results indicate that onconase appears to predominantly cleave miRNA precursors at UG and UU
residues, which are similar to the cleavage specificity of onconase observed in tRNAs17. In summary, our results indicate that onconase ubiquitously downregulates miRNA expression in
mesothelioma cells. Interestingly, our biochemical assays reveal that onconase preferentially degrades miRNA precursors and mildly affects mature forms of miRNAs. Given that miRNA precursors
resemble tRNAs, the well-documented downstream RNA species of onconase, with ∼70-nt long and hairpin structure, we speculate that an appropriate secondary structure might be required for
onconase substrates. Indeed, similar to the predominant cleavages of onconase in tRNAs located in the variable loop or stem of D-arm17, we found that the cleavage sites of onconase in miRNA
precursors are mapped at both loop and stem regions. Given that oncogenic miRNAs are upregulated in cancer cells while tumor suppressive miRNAs are often downregulated18, it is likely that
onconase exerts its antitumor activity through targeting miRNAs, i.e., mainly oncogenic miRNAs. Taken together, our study reveals miRNA precursors as a novel class of RNA targets for
onconase in addition to tRNAs, bringing new insights into the mechanisms of onconase-mediated cytotoxicity in cancer cells. REFERENCES * Pavlakis N, Vogelzang NJ . Ranpirnase--an antitumour
ribonuclease: its potential role in malignant mesothelioma. _Expert Opin Biol Ther_ 2006; 6:391–399. Article CAS Google Scholar * Lee JE, Raines RT . Ribonucleases as novel
chemotherapeutics: the ranpirnase example. _BioDrugs_ 2008; 22:53–58. Article CAS Google Scholar * Notomista E, Catanzano F, Graziano G, _et al_. Onconase: an unusually stable protein.
_Biochemistry_ 2000; 39:8711–8718. Article CAS Google Scholar * Haigis MC, Kurten EL, Raines RT . Ribonuclease inhibitor as an intracellular sentry. _Nucleic Acids Res_ 2003;
31:1024–1032. Article CAS Google Scholar * Dickson KA, Haigis MC, Raines RT . Ribonuclease inhibitor: structure and function. _Prog Nucleic Acid Res Mol Biol_ 2005; 80:349–374. Article
CAS Google Scholar * Juan G, Ardelt B, Li X, _et al_. G1 arrest of U937 cells by onconase is associated with suppression of cyclin D3 expression, induction of p16INK4A, p21WAF1/CIP1 and
p27KIP and decreased pRb phosphorylation. _Leukemia_ 1998; 12:1241–1248. Article CAS Google Scholar * Iordanov MS, Ryabinina OP, Wong J, _et al_. Molecular determinants of apoptosis
induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. _Cancer Res_ 2000; 60:1983–1994. CAS PubMed Google Scholar
* Altomare DA, Rybak SM, Pei J, _et al_. Onconase responsive genes in human mesothelioma cells: implications for an RNA damaging therapeutic agent. _BMC Cancer_ 2010; 10:34. Article CAS
Google Scholar * Goparaju CM, Blasberg JD, Volinia S, _et al_. Onconase mediated NFKβ downregulation in malignant pleural mesothelioma. _Oncogene_ 2011; 30:2767–2777. Article CAS Google
Scholar * Shen RL, Sun RL, Wang QC, Ou L, Fei J . Growth inhibition effect of onconase on B16 melanoma cells _in vivo_ and _in vitro_. _Chinese J Cell Biol_ 2007; 29: 901–904. CAS Google
Scholar * Jiang S, Zhang HW, Lu MH, _et al_. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. _Cancer Res_ 2010; 70:3119–3127.
Article CAS Google Scholar * Costinean S, Sandhu SK, Pedersen IM, _et al_. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by
miR-155 in B cells of Emicro-MiR-155 transgenic mice. _Blood_ 2009; 114:1374–1382. Article CAS Google Scholar * He M, Xu Z, Ding T, Kuang DM, Zheng L . MicroRNA-155 regulates
inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. _Cell Mol Immunol_ 2009; 6:343–352. Article CAS Google Scholar * Romania P, Lulli V, Pelosi E,
Biffoni M, Peschle C, Marziali G . MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. _Br J Haematol_ 2008;
143:570–580. CAS PubMed Google Scholar * Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T . MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human
hepatocellular cancer. _Gastroenterology_ 2007; 133:647–658. Article CAS Google Scholar * Lu Z, Liu M, Stribinskis V, _et al_. MicroRNA-21 promotes cell transformation by targeting the
programmed cell death 4 gene. _Oncogene_ 2008; 27:4373–4379. Article CAS Google Scholar * Suhasini AN, Sirdeshmukh R . Transfer RNA cleavages by onconase reveal unusual cleavage sites. _J
Biol Chem_ 2006; 281:12201–12209. Article CAS Google Scholar * Kasinski AL, Slack FJ . Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting
microRNAs for cancer therapy. _Nat Rev Cancer_ 2011; 11:849–864. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Prof En-Duo Wang (Institute of Biochemistry and
Cell Biology, SIBS, CAS) for providing human tRNAArg. This work was supported by grants from the Ministry of Science and Technology of China (2011CB811303, 2012CB910802, 2011CB966304) and
the National Natural Science Foundation of China (30970621, 31170754). AUTHOR INFORMATION Author notes * Meng Qiao and Li-Dong Zu: These two authors contributed equally to this work. AUTHORS
AND AFFILIATIONS * State Key Laboratory of Molecular Biology, Graduate School of Chinese Academy of Sciences, 200031, Shanghai, China Meng Qiao, Li-Dong Zu, Xiao-Hong He & Mo-Fang Liu *
Shanghai Key Laboratory of Molecular Andrology, 200031, Shanghai, China Meng Qiao, Li-Dong Zu, Xiao-Hong He & Mo-Fang Liu * Center for RNA Research, Institute of Biochemistry and Cell
Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 200031, Shanghai, China Meng Qiao, Li-Dong Zu, Xiao-Hong He & Mo-Fang Liu * Shanghai Research Center
for Model Organisms, 201210, Shanghai, China Ru-Ling Shen & Qing-Cheng Wang Authors * Meng Qiao View author publications You can also search for this author inPubMed Google Scholar *
Li-Dong Zu View author publications You can also search for this author inPubMed Google Scholar * Xiao-Hong He View author publications You can also search for this author inPubMed Google
Scholar * Ru-Ling Shen View author publications You can also search for this author inPubMed Google Scholar * Qing-Cheng Wang View author publications You can also search for this author
inPubMed Google Scholar * Mo-Fang Liu View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Mo-Fang Liu. ADDITIONAL
INFORMATION ( SUPPLEMENTARY INFORMATION is linked to the online version of the paper on the _Cell Research_ website.) SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION, FIGURE S1 Onconase
downregulates miR-155 and miR-21 and upregulates their targets in mesothelioma cells. (PDF 187 kb) SUPPLEMENTARY INFORMATION, FIGURE S2 Onconase ubiquitously downregulates miRNAs with high
abundance in mesothelioma cells. (PDF 145 kb) SUPPLEMENTARY INFORMATION, DATA S1 Materials and Methods (PDF 78 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Qiao, M., Zu, LD., He, XH. _et al._ Onconase downregulates microRNA expression through targeting microRNA precursors. _Cell Res_ 22, 1199–1202 (2012).
https://doi.org/10.1038/cr.2012.67 Download citation * Published: 24 April 2012 * Issue Date: July 2012 * DOI: https://doi.org/10.1038/cr.2012.67 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative