- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT A balance between cell proliferation and cell loss is essential for tumor progression. Although up to 90% of cells are lost in late-stage carcinomas, the progression and
characteristics of remnant living cells in tumor mass are unclear. Here we used molecular imaging to track the progression of living cells in a syngeneic tumor model, and _ex vivo_
investigated the properties of this population at late-stage tumor. The piggyBac transposon system was used to stably introduce the dual reporter genes, including monomeric red fluorescent
protein (mRFP) and herpes simplex virus type-1 thymidine kinase (HSV1-tk) genes for fluorescence-based and radionuclide-based imaging of tumor growth in small animals, respectively.
Iodine-123-labeled 5-iodo-2′-fluoro-1-beta-D-arabinofuranosyluracil was used as a radiotracer for HSV1-tk gene expression in tumors. The fluorescence- and radionuclide-based imaging using
the single-photon emission computed tomography/computed tomography revealed that the number of living cells reached the maximum at 1 week after implantation of 4T1 tumors, and gradually
decreased and clustered near the side of the body until 4 weeks accompanied by enlargement of tumor mass. The remnant living cells at late-stage tumor were isolated and investigated _ex
vivo_. The results showed that these living cells could form mammospheres and express cancer stem cell (CSC)-related biomarkers, including octamer-binding transcription factor 4, SRY
(sex-determining region Y)-box 2, and CD133 genes compared with those cultured _in vitro_. Furthermore, this HSV1-tk-expressing CSC-like population was sensitive to ganciclovir applied for
the suicide therapy. Taken together, the current data suggested that cells escaping from cell loss in late-stage tumors exhibit CSC-like characteristics, and HSV1-tk may be considered a
theranostic agent for targeting this population _in vivo_. SIMILAR CONTENT BEING VIEWED BY OTHERS QUANTITATIVE IN VIVO BIOLUMINESCENCE IMAGING OF ORTHOTOPIC PATIENT-DERIVED GLIOBLASTOMA
XENOGRAFTS Article Open access 21 September 2020 INHIBITION OF ORTHOTOPIC CASTRATION-RESISTANT PROSTATE CANCER GROWTH AND METASTASIS IN MICE BY JC VLPS CARRYING A SUICIDE GENE DRIVEN BY THE
PSA PROMOTER Article Open access 11 December 2023 WHOLE-BODY TRACKING OF SINGLE CELLS VIA POSITRON EMISSION TOMOGRAPHY Article 15 June 2020 MAIN Tumor growth is dependent on a kinetic model
that is based on the progression of cell proliferation and cell loss. The parameters for cell proliferation during tumor progression include the cell-cycle time (Tc), growth fraction (GF),
and potential tumor-doubling time (Tpot). In contrast, the cell-loss factor is determined by Tpot and the actual time for doubling of the tumor volume (Td).1 The causes of cell loss include
malnutrition and lack of oxygen caused by rapid proliferation, necrosis and apoptosis, immunological attack, escape from the primary site, and exfoliation.2 These conditions can be regarded
as stresses for cells residing in a rapidly growing tumor. Whether cells escaping from these stresses inherit or obtain resistance abilities is unknown. The tracking and characterization of
living cells in a tumor are important for cancer treatment. Reporter-gene imaging is an indirect approach to labeling cells for image-based _in vivo_ tracking and targeting by different
modalities.3 This method is especially important for tracking cell viability _in vivo_ because gene transcription and translation occur only in living cells.4 In addition, the transmission
of genes to progeny is in principle not diminished or diluted if the reporter genes can replicate within the genomes of host cells.5 Firefly luciferase and fluorescent proteins are canonical
reporter genes used for bioluminescent imaging and optical imaging, respectively. For radionuclide-based reporter-gene imaging, herpes simplex virus type-1 thymidine kinase (HSV1-tk) is
commonly used because it can uptake a broad range of radiolabeled nucleoside analogues by substrate phosphorylation for imaging the target cells _in vivo_.6 Expression of the HSV1-tk
reporter gene can be used for living cell tracking by positron emission tomography (PET) or single-photon emission computed tomography (SPECT), depending on the types of radionuclide-labeled
substrates. For instance, iodine-123-labeled 5-iodo-2′-fluoro-1-beta-D-arabinofuranosyluracil (123I-FIAU) is the most reliable radiolabeled nucleoside analogues for SPECT imaging of HSV1-tk
gene expression because it exhibits high tumor/background ratio _in vivo_.7, 8 As described above, the sensitivity and specificity of 123I-FIAU on cancer detection is based on the
expression of HSV1-tk reporter gene in the transduced cancer cells. These cancer cells can be distinguished from the non-cancer cells without HSV1-tk genes using the radionuclide imaging
modality. Technically, cultured cancer cells are stably transduced with HSV1-tk genes and implanted into animals for tumor formation. Using SPECT imaging, this cancer population can be
distinguished from non-cancer portion _in vivo_ by injecting 123I-FIAU as a radiotracer that are only accumulated in HSV1-tk gene expressing cancer cells.7 Multimodality reporter-gene
imaging using coexpressed luciferase/fluorescent proteins and HSV1-tk has been reported to be a powerful tool for basic biological and preclinical research.9, 10 In addition, PET and SPECT
can be merged with computed tomography (CT) to obtain functional/anatomic imaging with high sensitivity and spatial resolution. Although reporter-gene imaging is widely used for functional
studies _in vivo_, concerns regarding the safety and transduction pathways of exogenous genes hamper the clinical application of this approach. Viral-mediated methods are commonly used for
the transduction of reporter genes, but the uncontrolled infection and random genomic integration of genes of interest currently limit the clinical application of these methods.11 In
contrast, nonviral gene delivery is more acceptable in the clinic because biocompatible materials can be used, and the safety of these approaches is relatively easily assessed by
pharmacokinetic and pharmacodynamic studies.12 The piggyBac transposon system has recently attracted a great deal of attention because the system is a nonviral gene-delivery approach that
uses DNA-transposition ability for the stable expression of exogenous genes via a ‘cut-and-paste’ mechanism.13 The beauty of this system includes its known integration sites at ‘TTAA’
sequences and intron-preferred positions, mammalian compatibility, large cargo capacity, and its ability to be removed from the integration site without changing the DNA sequence.14 In
practice, this system requires a helper plasmid that encodes the piggyBac transposase gene to facilitate the transposition of exogenous gene.15 The piggyBac transposon system has been
applied in induced pluripotent stem cells, cancer research, and immunotherapy.16, 17 However, the use of this system has been less frequently reported in multimodality reporter-gene imaging
of tumor progression. Cancer stem cells (CSCs, or tumor-initiating cells) belong to the hierarchy model that a subset of rare cell population inherits stem cell-like characteristics,
including self-renewal and generation of non-tumorigenic progeny.18 This theory has intrigued many researchers in recent years because CSCs are resistant to chemoradiotherapy and are likely
to be the cause of tumor recurrence and metastasis.19 However, the identification of CSCs _in vivo_ remains a challenge because of the lack of suitable markers for this purpose. If CSCs
naturally resist environmental stresses, it would be speculated that this population may also escape from cell loss during tumor progression. More evidence is required to support this
hypothesis. The percentage of cell loss during tumor progression is approximately 40–90%, depending on the cancer type.1 The remnant viable cells may be important for promoting tumor growth
and metastasis. Reporter-gene imaging should be ideal to track these living cells for further investigation of their characteristics. In this study, we established a syngeneic tumor model
derived from 4T1 murine breast carcinomas transduced with monomeric red fluorescent protein (mRFP)/HSV1-tk dual reporter genes using the piggyBac transposon system. A combination of optical
imaging and SPECT/CT fusion imaging using 123I-FIAU as a probe was exploited to track the remnant living cells in late-stage primary tumors. Furthermore, _ex vivo_ studies showed that the
surviving cells exhibited CSC-like characteristics. These findings may contribute to therapeutic designs for cancer treatment. RESULTS ESTABLISHMENT OF THE TRANSGENIC TUMOR CELLS WITH
IMAGINABLE DUAL REPORTER GENES USING THE PIGGYBAC TRANSPOSON SYSTEM In this study, the piggyBac transposon system was used to establish stable cancer cell lines harboring mRFP and HSV1-tk
reporter genes that were driven by different promoters (Supplementary Figure 1A). The Act4-PBase helper plasmid and PB-2 R-puro donor plasmid were cotransfected in human 293 T cells, H1299
cells, MDA-MB-231 cells, and mouse 4T1 cells. The expression of reporter genes was sustained in each cell type for longer than 2 weeks, but cells transfected with donor plasmid only lost
reporter-gene expression in 4 days (Supplementary Figures 1B and C). To enrich pure stable cells for _in vivo_ imaging, we sorted the piggyBac-transfected cells expressing mRFP by
fluorescence-activated cell sorting (FACS). Here we focused on 4T1 cells because a syngeneic tumor model was subsequently established. Two days after transfection, 0.7–0.9% of
mRFP-expressing 4T1 cells could be sorted (Supplementary Figure 2). The expression of reporter genes was sustained in 4T1 cells cotransfected with both plasmids (named 4T1-PB-2R/PBase cells)
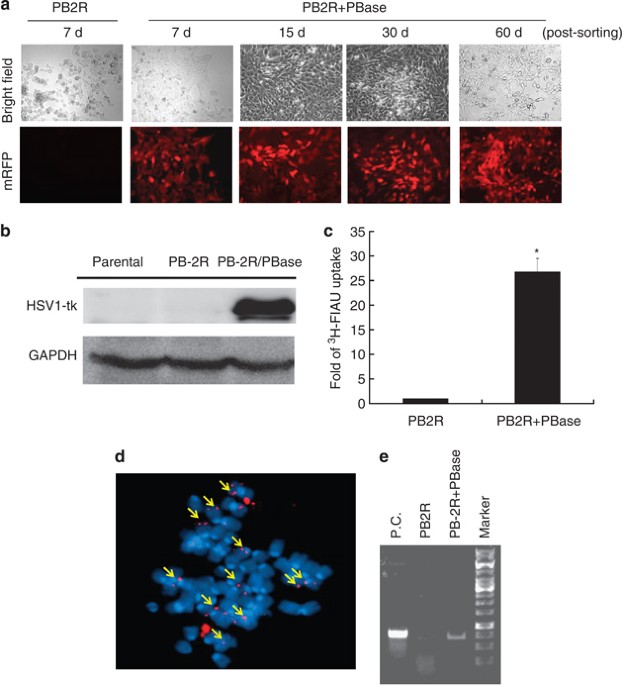
for up to 60 days without puromycin selection, but it was diminished in 4T1 cells transfected with PB-2 R-puro only (named 4T1-PB-2 R cells) after 7 days of culture (Figure 1a). The
transposition assay was also used to confirm the requirement for the Act-PBase helper plasmid in the stabilization of reporter genes in 4T1 cells by puromycin selection (Supplementary Figure
3). The sustained expression of HSV1-tk in 4T1-PB-2R/PBase cells was determined using the western blot assay (Figure 1b). HSV1-tk remained functional as determined by a cell-uptake assay
using 3H-FIAU (Figure 1c). Furthermore, 4T1-PB-2R/PBase cells, but not 4T1-PB-2R cells, were sensitive to ganciclovir (GCV), which is a prodrug phosphorylated and activated by HSV1-tk to
cause cell death (Supplementary Figure 4). Moreover, the fluorescence _in situ_ hybridization (FISH) assays showed that the long-term expression of reporter genes was associated with the
chromosomal integration of reporter genes in the genomes of 4T1-PB-2R/PBase cells (Figure 1d). This observation was also demonstrated using the genomic PCR (Figure 1e). This 4T1-PB-2R/PBase
cancer cell lines were subsequently used for fluorescence- and radionuclide-based imaging _in vivo_. IMAGING OF TUMOR FORMATION IN THE SYNGENEIC TUMOR MODEL To establish the syngeneic tumor
model, 4T1-PB-2R/PBase and 4T1-PB-2R tumor cells were subcutaneously implanted into BALB/c mice followed by imaging the mRFP expression. The growth rates of tumors formed by these two
transduced cell types and the untransduced parental 4T1 cells were similar, indicating that genomic integration of reporter genes did not affect cell proliferation (Figure 2a). Although both
transduced cell types formed tumors _in vivo_ within 7 days, the fluorescent signal was only detected in tumors formed by 4T1-PB-2R/PBase cells, but not by 4T1-PB-2R cells, within a cell
number-dependent manner (Figure 2b). The results of tumorous images formed by 4T1-PB-2R/PBase cells were quantified and compared among different cell amount after 1 day and 7 days of
subcutaneous (s.c.) implantation (Figure 2c). Furthermore, the histological investigation was used to demonstrate the coexpression of mRFP and HSV1-tk genes in this syngeneic tumor model
(Supplementary Figure 5). DETECTION OF THE REMNANT LIVING CELLS IN TUMOR PROGRESSION USING THE DUAL REPORTER-GENE IMAGING Rapid tumor growth is accompanied by an increase of cell loss at the
primary site. However, the characteristics of the remnant living cells in the tumor mass are less investigated. Hence, we determined to track the living 4T1-PB-2R/PBase cells during tumor
progression _in vivo_ based on the reporter-gene imaging. Figure 3a represented the agendas for imaging the expression of mRFP and HSV1-tk after s.c. implantation of 4T1-PB-2R/PBase breast
carcinomas (Figure 3a). For the fluorescence imaging, the mRFP signal at the tumor site was increased in the first 2 weeks, but was decreased thereafter (_n_=12) (Figure 3b). These results
suggest that the amount of viable cells in the primary tumor is reduced during tumor progression. To better understand the spatial and temporal characteristics of viable 4T1-PB-2R/PBase
cells in implanted tumors, mice were i.v. injected with 123I-FIAU to track cells expressing HSV1-tk gene using the microSPECT/CT. The results showed that the accumulation of 123I-FIAU in the
primary tumor site was detected on 7 days after implantation, but the ratio of accumulation tended to reduce during tumor progression (Figure 3c). This observation was further confirmed by
the reconstructed three-dimensional (3-D) imaging of living cells in the tumor mass (Figure 3c and Supplementary Video 6). The fluorescence and radionuclide imaging of tumors were
subsequently quantified by the selected region of interest (ROI) of each image. The amount of photon flux of fluorescence imaging was reduced after 11 days of tumor growth (Figure 3d). The
ROI signal of radionuclide imaging was also decreased after 7 days of tumor implantation, even though the tumor size was increased (Figure 3e). The imaging results of the fluorescent and
radionuclide images were also attempted to merge to display the quantificational correlations of living cell signal at the primary tumor (Supplementary Figure 7). Moreover, the
histopathological investigation were applied to confirm that functional tumor cells only occupied part of the late-stage tumor (Figure 3f). Apparent necrotic region was also detected.
Additionally, the anatomic examination of visceral showed that at least liver contained suspected tumor cells (Supplementary Figure 8A).They were subsequently confirmed by visualizing the
expression of mRFP as shown in the primary tumor formed by 4T1-PB-2R/PBase cells (Supplementary Figure 8B). REMNANT LIVING TUMOR CELLS EXHIBIT CSC-LIKE CHARACTERISTICS THROUGH _EX VIVO_
EXAMINATION Cell lost from rapidly growing tumors is caused by intrinsic and extrinsic stresses. It is speculated that cells escaping from these stresses may perform particular abilities to
survive and promote the tumor progression. Here we investigated whether the remnant living cells in the late-stage tumors exhibited the characteristics of CSCs, which are supposed to be
resistant to environmental stresses for tumor viability. Based on the results of the multimodality reporter-gene imaging described above, the _ex vivo_ experiments were performed to isolate
the remnant living 4T1-PB-2R/PBase cells from late-stage tumors formed in separate mice. First, the sphere-formation assay showed that mRFP-expressing mammospheres were formed in _in vivo_
tumor-isolated 4T1-PB-2R/PBase cells, but not in parental and original transduced 4T1 cells after seeding (Figure 4a). The number of mammospheres from _ex vivo_ 4T1-PB-2R/PBase cells was
greater than that of parental 4T1 cells after 4 days of initial seeding (Figure 4b). These observations suggest that remnant living cells in late-stage tumors exhibit increased
sphere-forming capacity, which is one of the CSC characteristics. Two general CSC biomarkers, octamer-binding transcription factor 4 (Oct4) and SRY (sex-determining region Y)-box 2 (Sox2),
were subsequently examined using semiquantitative real-time PCR (qPCR). Compared with parental 4T1 cells, _ex vivo_ 4T1-PB-2R/PBase cells exhibited upregulation of Oct4 and Sox2 mRNA
(Figures 5a and b). The transcriptional activity of Oct4 was also increased in _ex vivo_ 4T1-PB-2R/PBase cells in an Oct4 promoter assay (Figure 5c). Furthermore, the protein levels of Oct4,
Sox2 and another CSC-associated biomarkers CD133 were upregulated in _ex vivo_ 4T1-PB-2R/PBase cells isolated from late-stage tumors formed in different mice (Figure 5d). No significant
change of these biomarkers in original 4T1-PB-2R/PBase cells with routine subculture before _in vivo_ implantation. Taken together, the remnant living cells escaping from cell loss in
late-stage tumors display CSC-like characteristics, at least in part. HSV1-TK-EXPRESSING REMNANT LIVING TUMOR CELLS WITH CSC-LIKE CHARACTERISTICS ARE SENSITIVE TO GCV The expression of
HSV1-tk was also preserved in remnant 4T1-PB-2R/PBase living cells isolated from the late-stage tumor (Figure 6a). HSV1-tk is the first identified suicide gene that is considered safe for
gene therapy in transduced cancer cells after exposed to GCV.20 Given the remnant 4T1-PB-2R/PBase living cells isolated from the late-stage tumor _in vivo_ exhibited CSC-like
characteristics, we examined whether they remained sensitive to GCV. We first examined the sphere-formation capacity and side population percentage of original 4T1-PB-2R/PBase cells before
and after GCV to determine whether CSCs would be enriched by this treatment. The results showed that both CSC-related characteristics were not increased in GCV-treated 4T1-PB-2R/PBase cells,
suggesting that this method would not increase CSC population _in vitro_ (Supplementary Figure 9). Subsequently, the 3-(4,5-dimethylthiazol-2-yl)-2,5diphenylterazoliumbromide (MTT) assay
showed that the cell viability was greatly reduced in the isolated remnant 4T1-PB-2R/PBase living cells with CSC-like properties after they were exposed to different concentrations of GCV
(Figure 6b). Thus, HSV1-tk is not only important for reporter-gene imaging of CSC-like living cells in tumors but also a potent therapeutic agent in this cell population. DISCUSSION The
current data demonstrate that tumor cells escaping from cell loss and surviving in the late-stage tumors contain CSC-like characteristics. Conventionally, CSCs are believed to emerge after
the chemo- and radiotherapy because of the generation of resistance. Nevertheless, we found that cells would spontaneously express CSC-like characteristics when they were not lost in the
late-stage tumor. The reporter-gene imaging based on the optical and microSPECT/CT modalities is essential for tracking the spatial and temporal characteristics of these remnant living cells
_in vivo_. It would be of interest to further investigate if the stress of cell-loss factor can promote the generation of CSC-like population during tumor progression _in vivo_. CSCs have
been reported to be a rare population that is related to the recurrence and metastasis of advanced tumors.21 For experimental purposes, putative CSCs can be isolated from cultured cell lines
using sphere formation and FACS based on Hoechst 33342 dye exclusion or fluorescein-conjugated antibodies against the expression of stem cell markers.22 However, several lines of evidence
have argued that cells selected using these _in vitro_ methods exhibit negative or even opposite tumorigenic properties from those predicted by the CSC hypothesis.23, 24, 25 In addition, the
currently available markers for _in vitro_ CSC selection are not very suitable for _in vivo_ application.26 The lack of _in vivo_ evidence of CSCs would impede the clinical application of
this theory. Based on our _ex vivo_ experimental results, cells survived in late-stage tumor would exhibit CSC-like characteristics. Therefore, it suggests that remnant living cells in
advanced tumor are the CSCs _per se_. The reporter-gene imaging is likely the most appropriate approach to tracking these living tumor cells _in vivo_, at least in the preclinical study. The
reporter-gene imaging is based on the principle of molecular biology to track living cells _in vivo_. This approach is also appropriate for _in vivo_ imaging of CSCs.27 For instance, Vlashi
_et al._26 have established a short half-life version of the fluorescent ZsGreen reporter gene and demonstrated that CSCs contain reduced 26S proteasomal activity. They showed that a local
fractionated irradiation of tumor _in vivo_ steadily increased the ZsGreen-positive cells with CSC characteristics. In our study, we used constitutively expressed mRFP and HSV1-tk reporter
genes to track the remnant living cells in advanced tumor. Interestingly, after transfection of a short half-life version of green fluorescent protein (d2GFP) into these remnant living
cells, they also expressed sustained fluorescent signals compared with parental cells using the fluorescence microscopy (data not shown). Therefore, use of reporter-gene imaging to track the
remnant living cells in advanced tumor is likely a direct approach to find CSCs _in vivo_. The fluorescence imaging has been claimed to be ideal for _in vivo_ detection of the rare cell
population in tumors.28 Additionally, the radionuclide-based imaging is essential for tracking functional cells in deep tissues using the 3-D reconstruction. Indeed, mutant HSV1-tk has been
combined with firefly luciferase for imaging the differentiation of mesenchymal stem cells using PET and bioluminescence dual imaging modalities.9 Here we provide a proof-of-concept that a
similar dual reporter-gene imaging using mRFP and HSV1-tk can also track CSC-like viable cells in late-stage tumors using the syngeneic tumor model. These two reporter genes are driven by
separate promoters. This is important for tracking functional cells _in vivo_ using the multimodality imaging because reduction of both imaging signals in tumors should be due to cell loss
but not inactivation of one of the reporter genes. The current findings would encourage us to apply this approach to track the remnant living cells with CSC-like characteristics using the
xenograft tumor model in the future. The syngeneic tumor model is ideal for exploring the tumor growth and metastasis in the context of a normal immune system. This model is also especially
particularly important for the study of the growth niche for CSC progression.29 Bioluminescent reporter-gene imaging based on luciferase expression has been reported to track the growth and
metastasis of the 4T1 tumor model of late-stage breast cancer.30 Our 4T1-PB-2R/PBase tumor model contains a HSV1-tk reporter gene that can obtain better spatial resolution with 3-D images
for detecting tumor growth and potent metastasis using the microSPECT/CT. According to the _ex vivo_ investigation of mRFP expression, we were able to identify the 4T1-PB-2R/PBase cells that
existed not only in the primary site but also in the liver in late-stage tumor using the fluorescence microscope. Therefore, a multimodality-based reporter-gene system should be essential
for investigating the behaviors of living tumor cells in a syngeneic tumor model _in vivo_ and _ex vivo_. The theory of tumor kinetics was proposed approximately half a century ago.1, 31 The
cell-loss factor accounts for the fact that the measurable tumor growth rate is much slower than the prediction from the cell-cycle time of each individual cell and GF. In general, the
cell-loss factor tends to be larger (up to 90%) in carcinomas that usually exhibit apoptosis during progression.2 The remnant living cells in this tumor type are essential for determining
the efficacy of chemotherapy and radiotherapy because tumors with a high level of cell-loss factor respond quickly to these treatments. Characterization of these living cells should be
important for the design of therapeutic strategies. Because the current data suggest that these remnant living cells contain CSC-like characteristics, it would be possible to precisely track
these population using CSC-specific probe to identify their position and quantity in the future. CSCs have been reported to be a rare population that is related to the recurrence and
metastasis of advanced tumors.21 For experimental purposes, putative CSCs can be isolated from cultured cell lines using sphere formation and FACS based on Hoechst 33342 dye exclusion or
fluorescein-conjugated antibodies against the expression of stem cell markers.22 However, several lines of evidence have argued that cells selected using these _in vitro_ methods exhibit
negative or even opposite tumorigenic properties from those predicted by the CSC hypothesis.23, 24, 25 In addition, the currently available markers for _in vitro_ CSC selection are not very
suitable for _in vivo_ application.26 The lack of _in vivo_ evidence of CSCs would impede the clinical application of this theory. Based on our _ex vivo_ experimental results, cells survived
in late-stage tumor would exhibit CSC-like characteristics. Therefore, it suggests that remnant living cells in advanced tumor are the CSCs _per se_. The reporter-gene imaging is likely the
most appropriate approach to track these living tumor cells _in vivo_, at least in the preclinical study. Although CSCs have been regarded as potent targets for cancer therapy, these cells
usually exhibit resistance to chemotherapy and radiotherapy. As a reporter gene, we showed that the HSV1-tk expressed in the CSC-like population from 4T1 cells was susceptible to GCV,
suggesting that HSV1-tk-based suicide gene therapy may be effective in CSCs. These results are consistent with a previous report showing that lentiviral vector-mediated HSV1-tk gene therapy
can cause remission of cancer stem-like glioblastoma xenografts.32 Therefore, further investigation of whether the HSV1-tk gene can be used as a theranostic agent for targeting CSCs would be
of interest. In summary, we utilized reporter-gene imaging for _in vivo_ detection of living cells during tumor progression using the 4T1 syngeneic tumor model. For late-stage tumors, the
remnant living cells exhibit CSC-like characteristics, which may explain, at least in part, the tumor metastasis and chemoradiotherapeutic resistance that may cause the recurrence. Because
clinically detected human cancers have usually reached mid to late stages, the hypothesis that remnant living cells contain CSC-like characteristics will provide important information for
the design of novel therapeutic strategies for cancer treatment. MATERIALS AND METHODS CELL LINES, PLASMID AND STABLE TRANSFECTION Human embryonic kidney 293T cells (provided by National
RNAi Core Facility, Academia Sinica, Taipei, Taiwan, originated from American Type Culture Collection (ATCC no. CRL-11268)), human non-small lung cancer H1299 cells,33 and human breast
cancer MDA-MB-231 cells (a kind gift from Dr. Jeng-Jong Hwang at National Yang-Ming University, originated from ATCC no. HTB-26) were cultured in Dulbecco’s modified Eagle’s medium
supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamate, 50 U/ml penicillin and 50 _μ_g/ml streptomycin (Invitrogen Inc., Carlsbad, CA, USA). The cell lines were maintained at 37 °C
in a humidified incubator containing 5% CO2 and were routinely passaged every 2 days. 4T1 murine breast carcinomas (a kind gift from Dr. Yueh-Hsing Ou at National Yang-Ming University,
originated from ATCC no. CRL-2539), 4T1-PB-2R cells, and 4T1-PB-2R/PBase cells were cultured in RPMI1640 medium supplemented with 10% (v/v) FBS, 100 U/ml of penicillin, and 100 mg/ml
streptomycin. 4T1 breast carcinomas were isolated from the mammary tumor of BALB/c mouse according to the product description of ATCC. The PiggyBac transposon system including the PB-tk-mRFP
reporter plasmids and Act-PBase helper plasmid were kindly provided by Dr. Congjian Xu (Fudan University, People’s Republic of China).34 PB-tk-mRFP was further modified by inserting a
puromycin-resistance cassette into the _Bgl_II and _Bam_HI sites to obtain a new construct named PB-2R-puro. For cotransfection, PB-2R-puro was mixed with Act-PBase at the optimal ratio and
transfected into cells using jetPEI transfection reagent (Polyplus-Transfection Inc., New York, NY, USA). Transfected cells were sorted using a BD FACSCalibur system (BD, Franklin Lakes, NJ,
USA) equipped with an air-cooled argon laser excited at 488 nm. The cells were expanded and maintained in normal medium for further experiments. WESTERN BLOT ANALYSIS The procedure of
western blot analysis was performed as described previously.33 Anti-HSV1-tk (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), anti-Oct4, anti-Sox2 (Abcam, Cambridge, MA, USA), and
anti-CD133 (Cell Signaling Technology, Danvers, MA, USA) antibodies were used according to the conducted experiments. MEASUREMENT OF HSV1-TK ACTIVITY _IN VITRO_ The activity of HSV1-tk was
determined by a cell-uptake assay and MTT assay. For the cell-uptake assay, 1 × 105 cells were seeded in 24-well culture plates and cultured overnight. 3H-FIAU (1 _μ_Ci per well) was then
added to each well for 2 h. The radioactivities in the cells and in the supernatant were determined separately and normalized using a gamma counter (Wallac 1470 Wizard; Perkin Elmer,
Waltham, MA, USA). For the MTT assay, 800 cells were seeded in 96-well plates and cultured overnight. Different concentrations of GCV (Sigma-Aldrich, Inc., St. Louis, MO, USA) were added to
the wells and maintained for 4 days. MTT (1 mg/ml) (Sigma-Aldrich, Inc.) was then added to the cells for 4 h, dissolved in dimethyl sulfoxide, and measured at an absorbance of 570 nm on an
ELISA plate reader (Bio-Tek Instruments, Winooski, VT, USA). FLUORESCENCE _IN SITU_ HYBRIDIZATION The FISH analysis was performed according to previous reports.35 The PB-2R-puro plasmid was
labeled with biotin-16-dUTP using the nick translation (BioNick kit; Invitrogen Inc.). The fluorescein-conjugated avidin was used to detect biotin-16-dUTP-labeled PB-2R-puro hybridizing to
the chromosomes. GENOMIC PCR The genomic DNA was extracted using DNAzol (Invitrogen Inc.). The forward and reverse primers for amplifying the mRFP sequence were 5′-CCATGGGCTGGGAGGCCTCC-3′
and 5′-TTAACCCTAGAAAGATAGTCTG-3′, respectively. The PCR protocol was followed by the manufacturer’s instruction. MICE AND SYNGENEIC TUMOR MODEL The 6-week-old female BALB/c mice were
purchased from the Laboratory Animal Center, National Taiwan University College of Medicine (Taipei, Taiwan, ROC). For this study, 18 mice were used for s.c. tumor implantation and imaging
processes. The amount of mice used for different experiments has been described in the results. The animal use protocols have been reviewed and approved by the Institutional Animal Care and
Use Committee of National Yang-Ming University (approval number: 981225). Based on the experimental design, 4T1 cells and the derived stable cell lines expressing reporter genes were
implanted into each mouse at subcutaneously. The tumor volumes at s.c. positions were measured by caliper every 3 days and calculated using the following formula: Volume=Length (mm) × Width2
(mm2)/2. TUMOR IMAGING For the fluorescence imaging, an IVIS 50 system (Xenogen Inc., Alamda, CA, USA) was used to image the expression of mRFP in the tumors formed by 4T1-PB-2 R/PBase and
4T1-PB-2R cells. Mice were anesthetized using 2% isoflurane during the imaging process. The ROIs were acquired based on the signals emitted from the tumor positions and semiquantified as
photons per second. Data quantification was analyzed using the IGOR-PRO Living Imaging Software (WaveMetrics, Inc., Lake Oswego, OR, USA). MicroSPECT/CT tumor imaging was based on a FLEX
Triumph preclinical imaging system (Gamma Medica-Ideas, Inc., Northridge, CA, USA) as previously described.36 The Lugol’s solution (potassium iodide tablets) were given 1 h prior to i.v.
injection of 123I-FIAU (16 mCi/kg). Six hours later, the mice were anesthetized using 2% isoflurane mixed with oxygen and scanned by CT using 512 slides for anatomic coregistration.
Subsequently, a static SPECT sequence involving eight frames was conducted. Thirty-two projections (50 s) were acquired over 180°, which formed a 60 × 60 matrix for a total imaging time of
30 min per frame. The tumor images were viewed, reconstructed for 3-D images, and quantified using a free Amide software (SourceForge, Geeknet Inc., Fairfax, VA, USA).37 SPHERE-FORMATION
ASSAY Five-thousand cells were seeded in a 10-cm noncoated plate. The conditional medium contained serum-free DMEM/F12, 10 ng/ml of epithelial growth factor, 10 ng/ml of basic fibroblast
growth factor, 10 ng/ml of insulin, and 5 ml of N2 (Gibco Inc., Grand Island, NY, USA). Formed spheres were visualized under a bright-field microscope. REAL-TIME PCR ANALYSIS Total RNA was
extracted using TRIzol reagent (Invitrogen Inc.) and was purified using Direct-zol RNA miniprep kit (Zymo Research Corporation, Irvine, CA, USA) to remove genomic DNA contamination according
to the manufacturer’s instruction. An amount of 1 _μ_g of purified total RNA was subjected to cDNA synthesis using SuperScript II reverse transcriptase (Invitrogen Inc.). For qPCR analysis
of Oct4 gene expression, the forward primer was 5′-TTGGGCTAGAGAAGGATGTGGTT-3′, and the reverse primer was 5′-GGAAAAGGGACTGAGTAGAGTGTGG-3′. For Sox2 gene, the forward primer was
5′-GCACATGAACGGCTGGAGCAACG-3′, and the reverse primer was 5′-TGCTGCGAGTAGGACATGCTGTAGG-3′.38 For human/mouse _β_-actin gene, forward primer was 5′-GGAAATCGTGCGTGACATTAAG-3′, and the reverse
primer was 5′-GGCCATCTCTTGCTCGAAGT-3′. The experiments were conducted using the Master SYBR Green reagent mixed with cDNA templates and corresponding primers, and the reactions were
performed in a StepOnePlus machine (Applied Biosystems Inc., Carlsbad, CA, USA). The quantification of mRNA level and the melting curve analysis were completed by the bundled software
(StepOne software version. 2.1, Applied Biosystems Inc.). OCT4 PROMOTER ASSAY The PGL4.2-Oct4 luciferase reporter-gene construct containing the human Pou5f (Oct4) gene promoter was a gift
from Dr. Mu-Hwa Yang at National Yang-Ming University. This construct was transfected into cells for 48 h using jetPEI (Polyplus-Transfection Inc.) according to the manufacturer’s
instructions. The transfected cells were lysed and subjected to the luciferase assay using 50 mM of D-luciferin. The illuminant intensity was measured using the Wallac Victor 2 Multi-label
Counter (Perkin Elmer). Each datum was normalized to the total protein level. STATISTICAL ANALYSIS Experiments were independently conducted for the assessment of significant differences
between the control and experimental groups, and the results were represented by means±S.D. Significant differences were determined using Student’s _t_-test. Significantly different results
were defined as _P_<0.05. ABBREVIATIONS * CT: computed tomography * FBS: fetal bovine serum * mRFP: monomeric red fluorescent protein * HSV1-tk: herpes simplex virus type-1 thymidine
kinase * 123I-FIAU: iodine-123-labeled 5-iodo-2′-fluoro-1-beta-D-arabinofuranosyluracil * SPECT: single-photon emission computed tomography * PET: positron emission tomography * CSC: cancer
stem cell * Oct4: octamer-binding transcription factor 4 * Sox2: SRY (sex-determining region Y)-box 2 * GCV: ganciclovir * GF: growth fraction * ROI: region of interest * FISH: fluorescence
_in situ_ hybridization * qPCR: semiquantitative real-time PCR * FACS: fluorescence-activated cell sorting * MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5diphenylterazoliumbromide * 3-D:
three-dimensional REFERENCES * Steel GG . Cell loss as a factor in the growth rate of human tumours. _Eur J Cancer_ 1967; 3: 381–387. Article CAS PubMed Google Scholar * Cooper EH,
Bedford AJ, Kenny TE . Cell death in normal and malignant tissues. _Adv Cancer Res_ 1975; 21: 59–120. Article CAS PubMed Google Scholar * Serganova I, Mayer-Kukuck P, Huang R, Blasberg R
. Molecular imaging: reporter gene imaging. _Handb Exp Pharmacol_ 2008; 185/2: 167–223. Article Google Scholar * Acton PD, Zhou R . Imaging reporter genes for cell tracking with PET and
SPECT. _Q J Nucl Med Mol Imaging_ 2005; 49: 349–360. CAS PubMed Google Scholar * Kircher MF, Gambhir SS, Grimm J . Noninvasive cell-tracking methods. _Nat Rev Clin Oncol_ 2011; 8:
677–688. Article CAS PubMed Google Scholar * Diehn M, Clarke MF . Cancer stem cells and radiotherapy: new insights into tumor radioresistance. _J Natl Cancer Inst_ 2006; 98: 1755–1757.
Article PubMed Google Scholar * Wang HE, Yu HM, Liu RS, Lin M, Gelovani JG, Hwang JJ _et al_. Molecular imaging with 123I-FIAU, 18F-FUdR, 18F-FET, and 18F-FDG for monitoring herpes
simplex virus type 1 thymidine kinase and ganciclovir prodrug activation gene therapy of cancer. _J Nucl Med_ 2006; 47: 1161–1171. CAS PubMed Google Scholar * Haubner R, Avril N,
Hantzopoulos PA, Gansbacher B, Schwaiger M . _In vivo_ imaging of herpes simplex virus type 1 thymidine kinase gene expression: early kinetics of radiolabelled FIAU. _Eur J Nucl Med_ 2000;
27: 283–291. Article CAS PubMed Google Scholar * Love Z, Wang F, Dennis J, Awadallah A, Salem N, Lin Y _et al_. Imaging of mesenchymal stem cell transplant by bioluminescence and PET. _J
Nucl Med_ 2007; 48: 2011–2020. Article PubMed Google Scholar * Ray P . The pivotal role of multimodality reporter sensors in drug discovery: from cell based assays to real time molecular
imaging. _Curr Pharm Biotechnol_ 2011; 12: 539–546. Article CAS PubMed Google Scholar * De A, Lewis XZ, Gambhir SS . Noninvasive imaging of lentiviral-mediated reporter gene expression
in living mice. _Mol Ther_ 2003; 7: 681–691. Article CAS PubMed Google Scholar * Lam AP, Dean DA . Progress and prospects: nuclear import of nonviral vectors. _Gene Ther_ 2010; 17:
439–447. Article CAS PubMed PubMed Central Google Scholar * Claeys Bouuaert C, Chalmers RM . Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons.
_Genetica_ 2010; 138: 473–484. Article CAS PubMed Google Scholar * Kim A, Pyykko I . Size matters: versatile use of PiggyBac transposons as a genetic manipulation tool. _Mol Cell
Biochem_ 2011; 354: 301–309. Article CAS PubMed Google Scholar * Kang Y, Zhang X, Jiang W, Wu C, Chen C, Zheng Y _et al_. Tumor-directed gene therapy in mice using a composite nonviral
gene delivery system consisting of the piggyBac transposon and polyethylenimine. _BMC cancer_ 2009; 9: 126. Article PubMed PubMed Central Google Scholar * Nakazawa Y, Huye LE, Salsman
VS, Leen AM, Ahmed N, Rollins L _et al_. PiggyBac-mediated cancer immunotherapy using EBV-specific cytotoxic T-cells expressing HER2-specific chimeric antigen receptor. _Mol Ther_ 2011; 19:
2133–2143. Article CAS PubMed PubMed Central Google Scholar * VandenDriessche T, Ivics Z, Izsvak Z, Chuah MK . Emerging potential of transposons for gene therapy and generation of
induced pluripotent stem cells. _Blood_ 2009; 114: 1461–1468. Article CAS PubMed Google Scholar * Reya T, Morrison SJ, Clarke MF, Weissman IL . Stem cells, cancer, and cancer stem cells.
_Nature_ 2001; 414: 105–111. CAS PubMed Google Scholar * Zhang M, Rosen JM . Stem cells in the etiology and treatment of cancer. _Curr Opin Genet Dev_ 2006; 16: 60–64. Article PubMed
Google Scholar * Caruso M, Panis Y, Gagandeep S, Houssin D, Salzmann JL, Klatzmann D . Regression of established macroscopic liver metastases after in situ transduction of a suicide gene.
_Proc Natl Acad Sci USA_ 1993; 90: 7024–7028. Article CAS PubMed PubMed Central Google Scholar * Visvader JE, Lindeman GJ . Cancer stem cells in solid tumours: accumulating evidence and
unresolved questions. _Nat Rev Cancer_ 2008; 8: 755–768. Article CAS PubMed Google Scholar * Chiodi I, Belgiovine C, Dona F, Scovassi AI, Mondello C . Drug treatment of cancer cell
lines: a way to select for cancer stem cells? _Cancers_ 2011; 3: 1111–1128. Article CAS PubMed PubMed Central Google Scholar * Beier D, Hau P, Proescholdt M, Wischhusen J, Oefner PJ,
Aigner L _et al_. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. _Cancer Res_ 2007; 67: 4010–4015. Article CAS
PubMed Google Scholar * Broadley KW, Hunn MK, Farrand KJ, Price KM, Grasso C, Miller RJ _et al_. Side population is not necessary or sufficient for a cancer stem cell phenotype in
glioblastoma multiforme. _Stem Cells_ 2011; 29: 452–461. Article CAS PubMed Google Scholar * Schatton T, Frank MH . The _in vitro_ spheroid melanoma cell culture assay: cues on tumor
initiation? _J Invest Dermatol_ 2010; 130: 1769–1771. Article CAS PubMed PubMed Central Google Scholar * Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M _et al_. _In vivo_
imaging, tracking, and targeting of cancer stem cells. _J Natl Cancer Inst_ 2009; 101: 350–359. Article CAS PubMed PubMed Central Google Scholar * Hermann PC, Bhaskar S, Cioffi M,
Heeschen C . Cancer stem cells in solid tumors. _Sem Cancer Biol_ 2010; 20: 77–84. Article CAS Google Scholar * Hart LS, El-Deiry WS . Invincible, but not invisible: imaging approaches
toward _in vivo_ detection of cancer stem cells. _J Clin Oncol_ 2008; 26: 2901–2910. Article PubMed Google Scholar * Borovski T, De Sousa EMF, Vermeulen L, Medema JP . Cancer stem cell
niche: the place to be. _Cancer Res_ 2011; 71: 634–639. Article CAS PubMed Google Scholar * Tao K, Fang M, Alroy J, Sahagian GG . Imagable 4T1 model for the study of late stage breast
cancer. _BMC cancer_ 2008; 8: 228. Article PubMed PubMed Central Google Scholar * Frindel E, Malaise EP, Alpen E, Tubiana M . Kinetics of cell proliferation of an experimental tumor.
_Cancer Res_ 1967; 27: 1122–1131. CAS PubMed Google Scholar * Huszthy PC, Giroglou T, Tsinkalovsky O, Euskirchen P, Skaftnesmo KO, Bjerkvig R _et al_. Remission of invasive, cancer
stem-like glioblastoma xenografts using lentiviral vector-mediated suicide gene therapy. _PLoS One_ 2009; 4: e6314. Article PubMed PubMed Central Google Scholar * Tsai CH, Chiu SJ, Liu
CC, Sheu TJ, Hsieh CH, Keng PC _et al_. Regulated expression of cofilin and the consequent regulation of p27(kip1) are essential for G(1) phase progression. _Cell Cycle_ 2009; 8: 2365–2374.
Article CAS PubMed Google Scholar * Kang Y, Zhang XY, Jiang W, Wu CQ, Chen CM, Gu JR _et al_. The piggyBac transposon is an integrating non-viral gene transfer vector that enhances the
efficiency of GDEPT. _Cell Biol Int_ 2009; 33: 509–515. Article CAS PubMed Google Scholar * Dierlamm J, Wlodarska I, Michaux L, La Starza R, Zeller W, Mecucci C _et al_. Successful use
of the same slide for consecutive fluorescence in situ hybridization experiments. _Genes Chromosomes Cancer_ 1996; 16: 261–264. Article CAS PubMed Google Scholar * Yang FY, Wang HE, Lin
GL, Teng MC, Lin HH, Wong TT _et al_. Micro-SPECT/CT-based pharmacokinetic analysis of 99mTc-diethylenetriaminepentaacetic acid in rats with blood-brain barrier disruption induced by focused
ultrasound. _J Nucl Med_ 2011; 52: 478–484. Article CAS PubMed Google Scholar * Loening AM, Gambhir SS . AMIDE: a free software tool for multimodality medical image analysis. _Mol
Imaging_ 2003; 2: 131–137. Article PubMed Google Scholar * Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS _et al_. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the
Oct4/Sox2 complex in embryonic stem cells. _Mol Cell Biol_ 2005; 25: 6031–6046. Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank Dr. Yu
Kang and Dr. Congjian Xu (Fudan University, Shanghai, China) for providing Act-PBase and PB-tk-mRFP plasmids for this work. We thank Dr. Yann-Jang Chen for the technological support of the
FISH assay. We thank the facility supported by the Molecular and Genetic Imaging Core/National Research Program for Genomic Medicine at National Yang-Ming University. This study was
supported by National Science Council of Taiwan, ROC (NSC 99-2314-B-010-029-MY3; NSC 100-2627-M-010-001; and NSC 101-2623-E-010-002-NU) and a grant from the Ministry of Education, Aim for
the Top University Plan, National Yang-Ming University. AUTHOR INFORMATION Author notes * Y-L Chen and S-Y Wang: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS *
Department of Biomedical Imaging and Radiological Sciences, National Yang-Ming University, Taipei, Taiwan, ROC Y-L Chen, S-Y Wang, R-S Liu, H-E Wang, J-C Chen, C A Chang, L-T Lin, D T W Tan
& Y-J Lee * Department of Nuclear Medicine, National PET/Cyclotron Center, Taipei Veterans General Hospital, Taipei, Taiwan, ROC R-S Liu * Molecular and Genetic Imaging Core, National
Yang-Ming University Medical School, Taipei, Taiwan, ROC R-S Liu * Department of Medical Research and Education, Taipei Veterans General Hospital, Taipei, Taiwan, ROC S-H Chiou * Institute
of Pharmacology, National Yang-Ming University, Taipei, Taiwan, ROC S-H Chiou * Institute of Clinical Medicine, School of Medicine, National Yang-Ming University, Taipei, Taiwan, ROC S-H
Chiou * Biophotonics and Molecular Imaging Research Center (BMIRC), National Yang-Ming University, Taipei, Taiwan, ROC Y-J Lee Authors * Y-L Chen View author publications You can also search
for this author inPubMed Google Scholar * S-Y Wang View author publications You can also search for this author inPubMed Google Scholar * R-S Liu View author publications You can also
search for this author inPubMed Google Scholar * H-E Wang View author publications You can also search for this author inPubMed Google Scholar * J-C Chen View author publications You can
also search for this author inPubMed Google Scholar * S-H Chiou View author publications You can also search for this author inPubMed Google Scholar * C A Chang View author publications You
can also search for this author inPubMed Google Scholar * L-T Lin View author publications You can also search for this author inPubMed Google Scholar * D T W Tan View author publications
You can also search for this author inPubMed Google Scholar * Y-J Lee View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence
to Y-J Lee. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Edited by A Stephanou Supplementary Information accompanies the paper
on Cell Death and Disease website SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 (PDF 193 KB) SUPPLEMENTARY FIGURE 2 (PDF 181 KB) SUPPLEMENTARY FIGURE 3 (PDF 150 KB) SUPPLEMENTARY FIGURE
4 (PDF 100 KB) SUPPLEMENTARY FIGURE 5 (PDF 71 KB) SUPPLEMENTARY FIGURE 6 (PDF 121 KB) SUPPLEMENTARY FIGURE 7 (PDF 76 KB) SUPPLEMENTARY FIGURE 8 (PDF 249 KB) SUPP VIDEO 6 #1 (MPG 305 KB) SUPP
VIDEO 6 #2 (MPG 288 KB) SUPP VIDEO 6 #3 (MPG 338 KB) SUPP VIDEO 6 #4 (MPG 322 KB) RIGHTS AND PERMISSIONS This work is licensed under the Creative Commons Attribution-NonCommercial-No
Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Chen, YL., Wang, SY., Liu, RS. _et al._ Remnant living cells that escape cell loss in late-stage tumors exhibit cancer stem cell-like characteristics. _Cell Death Dis_ 3, e399 (2012).
https://doi.org/10.1038/cddis.2012.136 Download citation * Received: 19 July 2012 * Revised: 24 August 2012 * Accepted: 27 August 2012 * Published: 04 October 2012 * Issue Date: October 2012
* DOI: https://doi.org/10.1038/cddis.2012.136 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is
not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * cell-loss factor * remnant living cells * cancer
stem cells * syngeneic tumor model * piggyBac transposon system * reporter-gene imaging

:max_bytes(150000):strip_icc():focal(799x0:801x2)/kristen-bell1-2000-93f6684ca08c40a7b31b9716d073c51d.jpg)



