- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT BACKGROUND: The prognostic significance of BRAF and NRAS mutations in metastatic melanoma patients remains uncertain, with several studies reporting conflicting results, often
biased by the inclusion of patients treated with BRAF and MEK (MAPK) inhibitors. We therefore interrogated a historical cohort of patients free of the confounding influence of MAPK inhibitor
therapy. METHODS: Patients with available archival tissue first diagnosed with metastatic melanoma between 2002 and 2006 were analysed. Mutational analysis was performed using the OncoCarta
Panel. Patient characteristics, treatment outcome and survival were correlated with BRAF/NRAS mutation status. RESULTS: In 193 patients, 92 (48%) melanomas were BRAF-mutant, 39 (20%) were
NRAS-mutant and 62 (32%) were wild-type for BRAF/NRAS mutations (wt). There was no difference in response to chemotherapy based on mutation status (35–37%). The distant disease-free interval
(DDFI) was significantly shorter in patients with wt melanoma (27.9 months _vs_ 35.1 for BRAF and 49.1 for NRAS) although this was not significant in multivariate analysis. Survival from
stage IV melanoma diagnosis was not significantly different based on mutation status. The DDFI was significantly shorter in patients with BRAFV600K/R versus BRAFV600E melanoma in univariate
and multivariate analyses. CONCLUSIONS: BRAF and NRAS mutation status does not influence survival in metastatic melanoma. SIMILAR CONTENT BEING VIEWED BY OTHERS BRAFV600E-MUTANT METASTATIC
NSCLC: DISEASE OVERVIEW AND TREATMENT LANDSCAPE Article Open access 16 April 2024 EFFICACY OF PARP INHIBITOR THERAPY AFTER TARGETED BRAF/MEK FAILURE IN ADVANCED MELANOMA Article Open access
05 September 2024 CLINICAL AND GENOMIC CORRELATES OF IMATINIB RESPONSE IN MELANOMAS WITH KIT ALTERATIONS Article 23 August 2022 MAIN Activating mutations in the oncogenes _BRAF or NRAS_
occur in approximately 40 and 20% of melanomas, respectively, and result in constitutive activation of the mitogen-activated kinase (MAPK) cell signalling pathway (Davies et al, 2002; Platz
et al, 2008). Small molecule inhibitors of mutant BRAF and the downstream kinase MEK (MAPK inhibitors) have transformed the management of BRAF-mutant metastatic melanoma and improved overall
survival (OS) compared with standard chemotherapy in patients with _BRAF_V600 mutant metastatic melanoma (Chapman et al, 2011; Flaherty et al, 2012; Hauschild et al, 2012). Similarly,
although to a lesser extent, single agent MEK inhibition has shown activity in NRAS-mutant metastatic melanoma (Falchook et al, 2012; Ascierto et al, 2013), with a phase III trial currently
underway (NCT01763164). The presence of a _BRAF_ mutation in metastatic colorectal cancer is associated with a shorter OS compared with _KRAS_ mutant or _RAS/RAF_ wild-type disease (Van
Cutsem et al, 2011; Yokota et al, 2011; Toland et al, 2012). Similarly _BRAF_ mutations are associated with an increased risk of recurrence in papillary thyroid cancer (Elisei et al, 2012;
Prescott et al, 2012; Fernandez et al, 2013). The prognostic significance of a _BRAF_ mutation in metastatic melanoma is less clear. Recent analysis of survival in metastatic melanoma
patients were performed when BRAF and MEK inhibitors were available and some patients included received these therapies (Long et al, 2011; Jakob et al, 2012), making comparisons between the
_BRAF_-mutant and wild-type populations difficult. One study examining _BRAF_ status only (Long et al, 2011) reported no difference in survival from stage IV diagnosis between patients with
_BRAF_-mutant and wild-type metastatic melanoma; however, when the analysis was limited to patients with _BRAF_-mutant melanoma who did not receive a MAPK inhibitor, a significantly shorter
survival in BRAF-mutant patients was observed. It is unclear if this difference in survival was due to differences in the biology of _BRAF_-mutant versus wild-type melanoma or a selection
bias due to the non-random selection of _BRAF_-mutant patients for entry into the early phase clinical trials of MAPK inhibitors. Another study examining _BRAF_ and _NRAS_ status reported
that NRAS-mutant melanoma was associated with the poorest survival (Jakob et al, 2012). However, an earlier study found that _NRAS_-mutant melanoma was associated with improved survival
compared with _BRAF_-mutant or _BRAF/NRAS_ wild-type disease (Ugurel et al, 2007). This uncertainty regarding the prognostic significance of _BRAF_ and _NRAS_ mutations in metastatic
melanoma led us to perform a retrospective analysis in a cohort of patients with advanced melanoma who were treated before the availability of MAPK inhibitors. We sought to correlate _BRAF_
and _NRAS_ mutation status with clinicopathologic characteristics, response to chemotherapy and survival, as well as to determine the frequency of other oncogenic mutations in metastatic
melanoma. MATERIALS AND METHODS PATIENT SELECTION AND DATA COLLECTION This study was undertaken at the Melanoma Institute Australia (MIA) in conjunction with Westmead Hospital and Royal
Prince Alfred Hospital with human ethics review committee approval (Protocol No. X11-0023 and HREC/11/RPAH/32). All patients consented to data collection and enrolment in the melanoma
research database (MRD). Patients with newly diagnosed metastatic melanoma (stage IV) managed at MIA between 2002 and 2006 with available archival paraffin-embedded melanoma tissue suitable
for DNA extraction were included. To exclude the effect of survivor bias, which may occur at a quaternary referral cancer centre, patients not seen at the MIA before or within 4 weeks of
developing metastatic melanoma were excluded. Patient demographics, primary tumour characteristics (date of primary diagnosis, Breslow thickness, ulceration, mitotic rate, ulceration, N
stage), clinical details at the time of diagnosis of stage IV melanoma (M stage, serum lactate dehydrogenase (LDH), organ involvement), and data regarding progress after development of stage
IV disease (development of brain metastasis, treatment with systemic therapy and response to chemotherapy) were collected from the MRD and further review of the clinical record. For
patients with more than one primary melanoma, the ‘culprit’ primary deemed responsible for subsequent metastatic disease was designated using a previously described algorithm (Murali et al,
2012; Mann et al, 2013). Chemotherapy included dacarbazine, temzolomide, fotumustine, combined carboplatin and paclitaxel or experimental combinations including these agents. Immunotherapy
included vaccines and experimental agents. No patient was treated with IL-2, ipilimumab, class 1 BRAF inhibitors or MEK inhibitors. Treatment benefit was determined prospectively by the
clinician, with either disease stability or a reduction in tumour burden during treatment considered as a beneficial response. TUMOUR SAMPLES AND MOLECULAR TESTING Distant metastatic samples
were preferentially sampled over lymph nodes or primaries where available. DNA was extracted from one core sample taken from one archival formalin-fixed, paraffin-embedded (FFPE) tissue
block of melanoma for each patient in the study. DNA was extracted using NucleoSpin FFPE DNA Kit (Macherey Nagel, Düren, Germany) according to the manufacturer’s instruction with an
overnight proteinase digestion. The quality and quantity of the extracted DNA was assessed using NanoDrop ND-1000 Spectrophotometer. A minimum of 500 ng of DNA was required for successful
mutational analysis. All samples were successfully amplified and analysed for 238 variant targets in a 24 multiplex polymerase chain reactions (PCR) using the OncoCarta Panel v1.0 Kit
including 19 tumour-related genes such as _BRAF_, _NRAS_, _KIT_ and _PIK3CA_ (http://bioscience.sequenom.com/oncocarta-panel). The genotypes were called based on the matrix-assisted laser
desorption ionisation-time of flight mass spectrometry (MALDI-TOF) technology on the Sequenom MassArray platform. Specifically, the key targeted mutational hotspots in this assay were
G464R/V/E, G466R, F468C, G469A/E/R/S/V, D594V/G, F595L, G596R, L597Q/R/S/V, T599I, V600E/K/R/L, K601N/E for _BRAF_ and G12V/A/D/C/R/S, G13V/A/D/C/R/S, A18T, Q61L/R/P/H/E/H/K for _NRAS_.
STATISTICAL METHODS Clinical and pathologic features were tested for associations with _BRAF_ or _NRAS_ mutation status using simple cross-tabulations, independent samples _t_-test, Fisher’s
exact test, Pearson c2, and/or the Mann–Whitney _U_ test. The distant disease-free interval (DDFI) was measured from the date of culprit primary melanoma diagnosis to diagnosis of distant
metastatic disease. Overall survival was calculated from the date of diagnosis of stage IV melanoma to last follow-up (censored) or death from melanoma (event). Univariate survival analyses
was carried out using the Kaplan–Meier method together with the log-rank (Mantel–Cox) test to calculate statistical significance. Univariate hazard ratios (HRs), 95% confidence intervals
(95% CI), and corresponding _P_-values were obtained using Cox regression. A Bonferroni correction was applied to all _P_-values resulting from the univariate DDFI and survival analyses to
adjust for multiple comparisons. Multivariate survival analyses were conducted with Cox proportional hazards method. The proportionality assumption was inspected visually for each
categorical covariate. A two-tailed _P_-value of less than 0.05 was considered statistically significant. All analyses were prespecified and carried out with the IBM SPSS Statistic 19.0
software package. RESULTS PATIENTS, TUMOUR SAMPLES AND MUTATION FREQUENCY Between 2002 and 2006, 322 patients with a new diagnosis of metastatic melanoma were seen at MIA. Nine patients were
excluded because they were first diagnosed with metastatic disease more than 4 weeks before their first consultation at MIA. Archival FFPE melanoma tissue sufficient for DNA analysis was
available in 193 of the 313 eligible patients. Mutations were identified in tumours from 140 (73%) patients, and 10 (5%) patients had more than one mutation. _BRAF_ mutations were detected
in 92 patients (48%), and _NRAS_ mutations in 39 patients (20%) (Table 1). No targeted mutations were identified in 53 patients (27%). Of the patients with _BRAF_ mutations, 65 (71%) were
V600E and 18 (20%) were V600K. Of the patients with an _NRAS_ mutation, 33 (85%) were substitutions for glutamine at position 61 (Q61H/K/L/R) and 6 (15%) were substitutions for glycine at
amino acids 12 (G12C/D) or 13 (G13C/S). No tumours harboured both an _NRAS_ and _BRAF_ mutation. Twenty-three mutations, in 19 (10%) patients, were detected in genes other than _BRAF/NRAS_;
the most common were mutations in _KI_T (_n_=7, 4%) or _PIK3CA_ (_n_=7, 4%) (Supplementary Table S1). Correlations with clinical features (Supplementary Table S2) and survival analyses were
not performed based on mutations other than _BRAF_ or _NRAS_ genes because of the small numbers and the heterogeneity of the mutation types. Subsequent analyses were based on a patient’s
tumour BRAF and NRAS status, and three cohorts were compared and analysed: _BRAF_-mutant (_n_=92); _NRAS_-mutant (_n_=39); and those in whom no targeted mutation was found in _BRAF_ or
_NRAS_ (wt, _n_=62). PATIENT DEMOGRAPHICS AND CLINICOPATHOLOGIC FEATURES OF PRIMARY MELANOMA BASED ON BRAF AND NRAS MUTATION STATUS Patients with _BRAF_-mutant melanoma were significantly
younger at diagnosis of the culprit primary melanoma than those with wt melanoma (Median 53 versus 59 years, _P_=0.002) (Table 2). Acral lentiginous and demoplastic melanoma subtypes
appeared to be more common in the wt cohort; however, the small numbers in each subtype precluded statistical analysis (Table 2). There was no significant difference in Breslow thickness,
mitotic rate, presence of ulceration and nodal status between the three cohorts at diagnosis of the culprit primary melanoma (Table 2). CLINICAL CHARACTERISTICS AND TREATMENT RECEIVED FOR
STAGE IV MELANOMA Patients with _BRAF_-mutant disease were significantly younger than those with wt at first diagnosis of stage IV melanoma (median age 56 versus 63 years, _P_=0.03) (Table
3). A higher proportion of patients with _NRAS_-mutant melanoma had M1c (anatomically defined) melanoma at first diagnosis of stage IV compared with patients with BRAF-mutant or wt melanoma
(Table 3). Serum LDH at stage IV diagnosis was not associated with mutation status (Table 3). There was a trend to an increased incidence of liver and CNS metastasis at diagnosis of stage IV
disease in patients with _NRAS_-mutant melanoma, although the risk of developing CNS metastasis at any time was similar between the three groups (40–45%) (Table 3). The number of patients
who received systemic therapy, either chemotherapy or immunotherapy, was not different between the three cohorts (Table 3). There was no difference in clinician-assessed benefit from
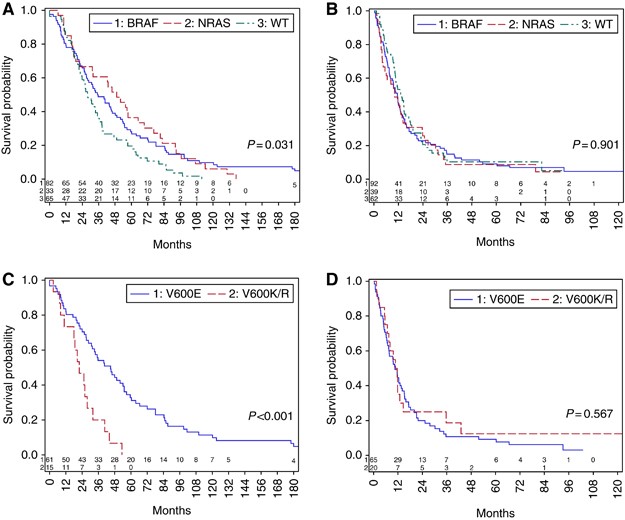
chemotherapy, (35–37%) (Supplementary Table S3). DISTANT DISEASE-FREE INTERVAL AND SURVIVAL ANALYSIS Although distant disease-free interval (DDFI) was significantly shorter in the wt cohort
(27.9 months) compared with either the _BRAF_ (35.1 months, _P_=0.03) or _NRAS_-mutant (median 49.1 months (_P_=0.01) populations (Figure 1A), this difference did not remain significant in
multivariate analysis when known prognostic factors for DDFI were included (Table 4). There was no difference in OS between the three cohorts from the time of diagnosis of stage IV melanoma
(Figure 1B) or in OS from culprit primary (Supplementary Figure S1). When analysed by _BRAF_ mutation genotype within the BRAF-mutant cohort, patients with _BRAF_V600K or R genotype melanoma
had a significantly shorter DDFI (_n_=20, median 22 months) than those with _BRAF_V600E (_n_=65, median 45 months, _P_=0.001) (Figure 1C); this difference remained significant in
multivariate analysis (Table 5). There was no difference in OS from diagnosis of stage IV disease between the _BRAF_V600K/R and the BRAFV600E patients (Figure 1D). There was a trend towards
a shorter survival from culprit primary in patients with _BRAF_V600K or R compared with _BRAF_V600E genotype melanoma (Supplementary Figure S2). The small numbers (_n_=7) of non-V600 _BRAF_
mutations precluded further analysis of this subgroup. There was a trend towards a shorter DDFI in patients who had an exon 2 (codon 61, _n_=23) compared with those with an exon 1 (codon
12/13, _n_=6) _NRAS_ mutation (Supplementary Figure S3, _P_=0.091). There was no difference in OS between the NRAS genotypes from diagnosis of stage IV disease (_P_=0.66). DISCUSSION This is
one of the largest studies to examine the prognostic significance of _BRAF_ and _NRAS_ mutation status in patients with metastatic melanoma, diagnosed and treated before the availability of
BRAF and MEK inhibitors. In contrast to other large studies, our survival analyses were not confounded by the availability BRAF and MEK inhibitors (Long et al, 2011; Jakob et al, 2012).
Previous large studies in primary melanomas were not powered to examine survival, as only 10% of patients with early-stage melanoma develop metastatic disease (Maldonado et al, 2003; Chang
et al, 2004; Houben et al, 2004; Shinozaki et al, 2004; Akslen et al, 2005), although one study found _NRAS_-mutant disease was associated with a poorer survival (Devitt et al, 2011a).
_NRAS_ mutations have also been found to be associated with fast growing primary melanomas (Nagore et al, 2013). In the metastatic melanoma population, the data regarding associations of
melanoma genotype and survival are conflicting (Edlundh-Rose et al, 2006; Ugurel et al, 2007; Long et al, 2011; Brissy et al, 2012; Jakob et al, 2012; Ekedahl et al, 2013). One study showed
that patients with _NRAS_-mutant tumours had an improved OS compared with those with _BRAF_-mutant or wt tumours (Ugurel et al, 2007), whereas another study suggested _NRAS_-mutant melanoma
predicted a poorer OS from stage IV disease (Jakob et al, 2012). A further study found that the presence of either _NRA_S or _BRAF_ mutations was associated with a poorer survival in the
setting of metastatic disease (Houben et al, 2004). Our finding that mutation status is not prognostic in the setting of stage IV melanoma is in keeping with other studies before the
availability of BRAF and MEK inhibitors (Chang et al, 2004; Edlundh-Rose et al, 2006). The data regarding the prognostic impact of mutation status in patients with stage III disease are
similarly conflicting. Some studies found no prognostic impact of mutation status (Rutkowski et al, 2012) and others found an association between _BRAF_-mutant melanoma and poorer OS (Moreau
et al, 2012; Mann et al, 2013). Although we found a significantly shorter DDFI in patients with wt disease, which has not been shown in prior studies that tested both _BRAF_ and _NRAS_
mutations (Jakob et al, 2012), it was not significant in multivariate analysis suggesting important differences in prognostic variables in the culprit primary melanoma. There are many
possible reasons for the lack of consistent results regarding the prognostic impact of _BRAF_ and _NRAS_ mutation status in both early and advanced melanoma, including the mutation testing
method, patient selection and geographic variations in the risk of specific melanoma mutations. Different mutation testing methodologies with different sensitivities and specificities were
used in various studies. In contrast to our study, very few prior studies analysed for a comprehensive range of melanoma-associated _BRAF_ and _NRAS_ mutations. As an example, a subset of
studies did not test for exon 1 (codon 12/13) _NRAS_ mutations (Edlundh-Rose et al, 2006; Devitt et al, 2011a) whereas others limited survival analysis only to _BRAF_ substitutions for
valine at codon 600 (Jakob et al, 2012). The OncoCarta assay is robust for FFPE samples and sensitive (detection limit of 10%) for the targeted hotspots within _BRAF_ and _NRAS_. Although
this assay does not analyse the complete genes or exons of interest, it does include all the key melanoma-associated _NRAS_ mutations in exons 1 and 2 and _BRAF_ mutations in exons 11 and 15
(Greaves et al, 2012). Patient selection varied substantially between studies, and may be the most important factor influencing the different results between them. Methods of patient
selection include selection of consecutive patients (Long et al, 2011; Moreau et al, 2012), selecting patients from a clinical database (Jakob et al, 2012), or, as in this study, selecting
patients on the basis of available tissue (Houben et al, 2004; Rutkowski et al, 2012; Mann et al, 2013). This study minimised other selection biases by including all patients seen within a
defined time period and limited the effect of survival bias by excluding those referred to our clinical service more than 4 weeks after the first diagnosis of metastatic melanoma. Each
method of patient selection is associated with potential biases; clinically accrued data sets are likely to enhance for survivors, particularly as initially testing was performed for entry
onto clinical trials or access to novel therapies, with a consequent referral bias of healthier and fitter patients who are willing to travel for experimental treatments (Long et al, 2011;
Jakob et al, 2012). Studies in which patients are selected based on available archival tissue may skew the population towards patients who have had surgery for stage III or IV disease, who
may represent a separate prognostic group with different mutational profiles. Geographical variations may also explain the differences in the impact of mutation status on prognosis.
_BRAF__V600K_ mutation is associated with chronic UV damage (Menzies et al, 2012) and varies by geography (Houben et al, 2004; Edlundh-Rose et al, 2006; Ugurel et al, 2007; Long et al, 2011;
Amanuel et al, 2012; Jakob et al, 2012; Menzies et al, 2012). Little is known about other genetic or epigenetic factors, which can occur concurrently with _BRAF_ and _NRAS_ mutations, and
may vary between regions, with possible prognostic implications, for example, PTEN loss is uncommon in NRAS-mutant melanoma but occurs in BRAF-mutant melanoma and can activate the PI3K
pathway (Hodis et al, 2012). Although NRAS mutations cause both PI3K/AKT and MAPK pathway activation (Tsao et al, 2000), it remains to be determined if activation of the PI3K pathway is
prognostic in melanoma. This dual pathway activation is one hypothesis to explain the association between NRAS mutations and a poorer prognosis compared with BRAF mutations in the previous
studies of melanoma (Jakob et al, 2012). However, this is not the case in colorectal cancer where BRAF mutations carry a poorer prognosis compared with KRAS mutant disease (Van Cutsem et al,
2011; Yokota et al, 2011; Toland et al, 2012). A meta-analysis may help to clarify the effect of mutation status on survival. We found no differences in the clinicopathologic factors of the
antecedent primary melanoma based on the mutation status, similar to prior studies (Shinozaki et al, 2004; Edlundh-Rose et al, 2006), but in contrast to one study which reported association
between _BRAF_ positivity and thinner primaries with lower numbers of mitoses (Devitt et al, 2011b). There was no association between the patterns of organ involvement of metastatic disease
both at the time of distant metastasis (stage IV) diagnosis and _BRAF/NRAS_ mutation status, although we found non-lung visceral metastases (M1c disease) more common in patients with
_NRAS_-mutant disease. One previous study reported an association between _BRAF_ or _NRAS_ mutations and the presence of CNS metastases at first occurrence of stage IV disease (Jakob et al,
2012). Our data show a trend towards higher rates of brain metastasis at initial stage IV diagnosis in keeping with this, but we show for the first time that the risk of developing brain
metastasis at any time is comparable at 40–45% of patients irrespective of _BRAF/NRAS_ mutation status. The response to chemotherapy was not influenced by mutation status. Although _BRAF_
wild-type melanomas have been reported to have a higher response rate to regional chemotherapy (Gallagher et al, 2008), in keeping with our data, mutation status did not influence response
or survival to systemic therapy with nab-paclitaxel or dacarbazine (Hersh et al, 2013). Exploratory analysis have found an association between _BRAF_ wild-type tumours and response to an
investigational combination including the anti-angiogenic therapy bevacizumab (von Moos et al, 2011), _NRAS_ mutations have also been reported to be associated with an improved response to
immunotherapy as compared with patients having _BRAF/NRAS_ wild-type tumours (Johnson et al, 2013). Analysis as part of future clinical trials should determine if mutation status is
predictive in treatments beyond those involving inhibitors of the MAPK pathway. Our finding that tumours with a _BRA_FV600K/R mutation have a significantly shorter DDFI than those with the
more common BRAFV600E mutation confirms prior reports (Menzies et al, 2012; Bucheit et al, 2013). We found no impact of _BRAF_ mutation genotype on OS from the time of first diagnosis of
stage IV melanoma, as previously reported in a different cohort from our institution (Menzies et al, 2012), although another study found V600K mutations were associated with a poorer
survival from stage IV (Bucheit et al, 2013). Analysis of DDFI and survival from stage IV diagnosis based on exon 1 or 2 _NRAS_ mutations showed no significant difference based on genotype,
in keeping with prior studies (Bucheit et al, 2013). Nevertheless, this exploratory analysis is limited by the relatively small number of patients with _NRAS_ mutations in this cohort. In
conclusion, our data show that _BRAF_ and _NRAS_ mutations are not prognostic in advanced melanoma. We confirmed the association between the _BRAF_V600K/R genotype and a shorter DDFI
compared with _BRAF_V600E in an independent cohort, but there was no difference in survival from stage IV diagnosis. Given the activity of BRAF and MEK inhibitors, future studies examining
the prognostic impact of _BRAF_ mutation status in advanced melanoma will be difficult to interpret and future studies should examine other genetic factors which may explain the conflicting
results seen across multiple studies. CHANGE HISTORY * _ 15 JULY 2014 This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at
publication _ REFERENCES * Akslen LA, Angelini S, Straume O, Bachmann IM, Molven A, Hemminki K, Kumar R (2005) BRAF and NRAS mutations are frequent in nodular melanoma but are not associated
with tumor cell proliferation or patient survival. _J Invest Dermatol_ 125 (2): 312–317. Article CAS PubMed Google Scholar * Amanuel B, Grieu F, Kular J, Millward M, Iacopetta B (2012)
Incidence of BRAF p.Val600Glu and p.Val600Lys mutations in a consecutive series of 183 metastatic melanoma patients from a high incidence region. _Pathology_ 44 (4): 357–359. Article CAS
PubMed Google Scholar * Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CM, Queirolo P, Blank CU, Hauschild A, Beck JT, St-Pierre A, Niazi F, Wandel S, Peters M, Zubel A,
Dummer R (2013) MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. _Lancet Oncol_ 14 (3): 249–256. Article CAS
PubMed Google Scholar * Brissy S, Gaudy-Marqueste C, Mallet S, Monestier S, Hesse S, Koeppel M-C, Rojat-Habib M-C, Nanni I, Loundou A, Lh Ouafik, Bonnet N, Richard M-A, Grob JJ (2012)
BRAF mutation as a pejorative marker in metastatic melanoma. _J Clin Oncol_ 30 (suppl): abstr 8555. Google Scholar * Bucheit AD, Syklawer E, Jakob JA, Bassett RL Jr, Curry JL, Gershenwald
JE, Kim KB, Hwu P, Lazar AJ, Davies MA (2013) Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. _Cancer_ 119 (21): 3821–3829.
Article CAS PubMed Google Scholar * Chang DZ, Panageas KS, Osman I, Polsky D, Busam K, Chapman PB (2004) Clinical significance of BRAF mutations in metastatic melanoma. _J Transl Med_ 2
(1): 46. Article PubMed PubMed Central Google Scholar * Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe
C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AMM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur AG (2011) Improved survival
with vemurafenib in melanoma with BRAF V600E mutation. _N Engl J Med_ 364 (26): 2507–2516. Article CAS PubMed PubMed Central Google Scholar * Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C,
Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A,
Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in
human cancer. _Nature_ 417 (6892): 949–954. Article CAS PubMed Google Scholar * Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen C-Y, Dobrovic A, McArthur G (2011a) Clinical outcome
and pathological features associated with NRAS mutation in cutaneous melanoma. _Pigment Cell Melanoma Res_ 24 (4): 666–672. Article CAS PubMed Google Scholar * Devitt B, Liu W, Salemi R,
Wolfe R, Kelly J, Tzen CY, Dobrovic A, McArthur G (2011b) Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. _Pigment Cell Melanoma Res_ 24 (4):
666–672. Article CAS PubMed Google Scholar * Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, Lundeberg J (2006) NRAS and BRAF mutations in melanoma tumours in
relation to clinical characteristics: a study based on mutation screening by pyrosequencing. _Melanoma Res_ 16 (6): 471–478. Article CAS PubMed Google Scholar * Ekedahl H, Cirenajwis H,
Harbst K, Carneiro A, Nielsen K, Olsson H, Lundgren L, Ingvar C, Jonsson G (2013) The clinical significance of BRAF and NRAS mutations in a clinic-based metastatic melanoma cohort. _Br J
Dermatol_ 169 (5): 1049–1055. Article CAS PubMed Google Scholar * Elisei R, Viola D, Torregrossa L, Giannini R, Romei C, Ugolini C, Molinaro E, Agate L, Biagini A, Lupi C, Valerio L,
Materazzi G, Miccoli P, Piaggi P, Pinchera A, Vitti P, Basolo F (2012) The BRAF(V600E) mutation is an independent, poor prognostic factor for the outcome of patients with low-risk
intrathyroid papillary thyroid carcinoma: single-institution results from a large cohort study. _J Clin Endocrinol Metab_ 97 (12): 4390–4398. Article CAS PubMed Google Scholar * Falchook
GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, Sun P, Moy C, Szabo SA, Roadcap LT, Peddareddigari VG, Lebowitz PF, Le NT, Burris HA 3rd, Messersmith WA, O'Dwyer PJ,
Kim KB, Flaherty K, Bendell JC, Gonzalez R, Kurzrock R, Fecher LA (2012) Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial.
_Lancet Oncol_ 13 (8): 782–789. Article CAS PubMed PubMed Central Google Scholar * Fernandez IJ, Piccin O, Sciascia S, Cavicchi O, Repaci A, Vicennati V, Fiorentino M (2013) Clinical
significance of BRAF mutation in thyroid papillary cancer. _Otolaryngol Head Neck Surg_ 148 (6): 919–925. Article PubMed Google Scholar * Flaherty KT, Robert C, Hersey P, Nathan P, Garbe
C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, Dummer R, Trefzer U, Larkin JMG, Utikal J, Dreno B, Nyakas M, Middleton MR, Becker JC, Casey M, Sherman LJ, Wu FS, Ouellet D, Martin
A-M, Patel K, Schadendorf D (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. _N Engl J Med_ 367 (2): 107–114. Article CAS PubMed Google Scholar * Gallagher SJ,
Thompson JF, Indsto J, Scurr LL, Lett M, Gao BF, Dunleavey R, Mann GJ, Kefford RF, Rizos H, Gallagher SJ, Thompson JF, Indsto J, Scurr LL, Lett M, Gao B-F, Dunleavey R, Mann GJ, Kefford RF,
Rizos H (2008) p16INK4a expression and absence of activated B-RAF are independent predictors of chemosensitivity in melanoma tumors. _Neoplasia_ 10 (11): 1231–1239. Article CAS PubMed
PubMed Central Google Scholar * Greaves WO, Verma S, Patel KP, Davies MA, Barkoh BA, Galbincea JM, Yao H, Lazar A, Aldape KD, Medeiros LJ, Luthra R (2012) Frequency and spectrum of BRAF
mutations in a retrospective, single-institution study of 1112 cases of melanoma. _J Mol Diagn_ 26 (12): 00310–00318. Google Scholar * Hauschild A, Grob J-J, Demidov LV, Jouary T, Gutzmer
R, Millward M, Rutkowski P, Blank CU, Miller WH, Kaempgen E, Martín-Algarra S, Karaszewska B, Mauch C, Chiarion-Sileni V, Martin A-M, Swann S, Haney P, Mirakhur B, Guckert ME, Goodman V,
Chapman PB (2012) Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. _Lancet_ 380 (9839): 358–365. Article CAS PubMed Google
Scholar * Hersh E, Del Vecchio M, Brown MP, Kefford R, Loquai C, Testori A, Robert C, Li M, Elias I, Renschler MF, Hauschild A (2013) A phase III trial of nab-paclitaxel versus dacarbazine
in chemotherapy-naive patients with metastatic melanoma: a subanalysis based on BRAF status. _J Clin Oncol_ 31 (suppl): abstr 9030. Google Scholar * Hodis E, Watson Ian R, Kryukov Gregory
V, Arold Stefan T, Imielinski M, Theurillat J-P, Nickerson E, Auclair D, Li L, Place C, DiCara D, Ramos Alex H, Lawrence Michael S, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N,
Onofrio Robert C, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton Donald L, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury John E, Davies Michael A, Gershenwald Jeffrey E,
Wagner Stephan N, Hoon Dave SB, Schadendorf D, Lander Eric S, Gabriel Stacey B, Getz G, Garraway Levi A, Chin L (2012) A landscape of driver mutations in melanoma. _Cell_ 150 (2): 251–263.
Article CAS PubMed PubMed Central Google Scholar * Houben R, Becker JC, Kappel A, Terheyden P, Brocker EB, Goetz R, Rapp UR (2004) Constitutive activation of the Ras-Raf signaling
pathway in metastatic melanoma is associated with poor prognosis. _J Carcinog_ 3 (1): 6. Article PubMed PubMed Central Google Scholar * Jakob JA, Bassett RL Jr, Ng CS, Curry JL, Joseph
RW, Alvarado GC, Rohlfs ML, Richard J, Gershenwald JE, Kim KB, Lazar AJ, Hwu P, Davies MA (2012) NRAS mutation status is an independent prognostic factor in metastatic melanoma. _Cancer_ 118
(16): 4014–4023. Article CAS PubMed Google Scholar * Johnson DB, Lovly CM, Flavin M, Ayers GD, Zhao Z, Iams WT, Iafrate AJ, Berry EG, Terry CR, Sullivan RJ, Carvajal RD, Sosman JA
(2013) NRAS mutation: A potential biomarker of clinical response to immune-based therapies in metastatic melanoma (MM). _J Clin Oncol_ 31 (suppl): abstr 9019. Google Scholar * Long GV,
Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF (2011) Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic
melanoma. _J Clin Oncol_ 29 (10): 1239–1246. Article PubMed Google Scholar * Maldonado JL, Fridlyand J, Patel H, Jain AN, Busam K, Kageshita T, Ono T, Albertson DG, Pinkel D, Bastian BC
(2003) Determinants of BRAF mutations in primary melanomas. _J Natl Cancer Inst_ 95 (24): 1878–1890. Article CAS PubMed Google Scholar * Mann GJ, Pupo GM, Campain AE, Carter CD, Schramm
SJ, Pianova S, Gerega SK, De Silva C, Lai K, Wilmott JS, Synnott M, Hersey P, Kefford RF, Thompson JF, Yang YH, Scolyer RA (2013) BRAF mutation, NRAS mutation, and the absence of an
immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. _J Invest Dermatol_ 133 (2): 509–517. Article CAS PubMed Google Scholar * Menzies AM,
Haydu LE, Visintin L, Carlino MS, Howle JR, Thompson JF, Kefford RF, Scolyer RA, Long GV (2012) Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant
metastatic melanoma. _Clin Cancer Res_ 18 (12): 3242–3249. Article CAS PubMed Google Scholar * Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Helias-Rodzewicz Z, Marin C, Peschaud
F, Chagnon S, Zimmermann U, Clerici T, Emile JF (2012) Prognostic value of BRAF(V(6)(0)(0)) mutations in melanoma patients after resection of metastatic lymph nodes. _Ann Surg Oncol_ 19
(13): 4314–4321. Article PubMed Google Scholar * Murali R, Brown PT, Kefford RF, Scolyer RA, Thompson JF, Atkins MB, Long GV (2012) Number of primary melanomas is an independent predictor
of survival in patients with metastatic melanoma. _Cancer_ 118 (18): 4519–4529. Article PubMed Google Scholar * Nagore E, Hacker E, Martorell-Calatayud A, Traves V, Guillen C, Hayward
NK, Whiteman D (2013) Prevalence of BRAF and NRAS mutations in fast-growing melanomas. _Pigment Cell Melanoma Res_ 26 (3): 429–431. Article CAS PubMed Google Scholar * Platz A, Egyhazi
S, Ringborg U, Hansson J (2008) Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. _Mol Oncol_ 1 (4): 395–405.
Article PubMed Google Scholar * Prescott JD, Sadow PM, Hodin RA, Le LP, Gaz RD, Randolph GW, Stephen AE, Parangi S, Daniels GH, Lubitz CC (2012) BRAF V600E status adds incremental value
to current risk classification systems in predicting papillary thyroid carcinoma recurrence. _Surgery_ 152 (6): 984–990. Article PubMed Google Scholar * Rutkowski P, Jurkowska M, Gos A,
Tysarowski A, Michej W, Switaj T, Dziewirski W, Zdzienicki M, Falkowski S, Olszewski WT, Siedlecki JA (2012) Correlations of molecular alterations in clinical stage III cutaneous melanoma
with clinical-pathological features and patients outcome. _J Clin Oncol_ 30 (suppl): abstr 8548. Google Scholar * Shinozaki M, Fujimoto A, Morton DL, Hoon DS (2004) Incidence of BRAF
oncogene mutation and clinical relevance for primary cutaneous melanomas. _Clin Cancer Res_ 10 (5): 1753–1757. Article CAS PubMed Google Scholar * Toland AE, Safaee Ardekani G,
Jafarnejad SM, Tan L, Saeedi A, Li G (2012) The prognostic value of BRAF mutation in colorectal cancer and melanoma: a systematic review and meta-analysis. _PLoS One_ 7 (10): e47054. Article
Google Scholar * Tsao H, Zhang X, Fowlkes K, Haluska FG (2000) Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. _Cancer Res_ 60 (7): 1800–1804.
CAS PubMed Google Scholar * Ugurel S, Thirumaran RK, Bloethner S, Gast A, Sucker A, Mueller-Berghaus J, Rittgen W, Hemminki K, Becker JC, Kumar R, Schadendorf D (2007) B-RAF and N-RAS
mutations are preserved during short time _in vitro_ propagation and differentially impact prognosis. _PLoS One_ 2 (2): e236. Article PubMed PubMed Central Google Scholar * Van Cutsem E,
Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F (2011) Cetuximab plus
irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. _J
Clin Oncol_ 29 (15): 2011–2019. Article CAS PubMed Google Scholar * von Moos R, Seifert B, Simcock M, Goldinger SM, Gillessen S, Ochsenbein A, Michielin O, Cathomas R, Schlappi M, Moch
H, Schraml PH, Mjhic-Probst D, Mamot C, Schonewolf N, Dummer R (2011) First-line temozolomide combined with bevacizumab in metastatic melanoma: a multicentre phase II trial (SAKK 50/07).
_Ann Oncol_ 23 (2): 531–536. Article PubMed Google Scholar * Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, Yatabe Y (2011) BRAF
mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. _Br J Cancer_ 104 (5): 856–862. Article CAS PubMed PubMed Central Google Scholar Download
references ACKNOWLEDGEMENTS This work was supported by Program Grant 633004 and project grants of the National Health and Medical Research Council of Australia (NHMRC), and an educational
grant from Roche Products, Pty Limited (Australia). RA Scolyer, GV Long and SA O’Toole are recipients of Cancer Institute New South Wales, Research Fellowships and RA Scolyer is supported by
an NHMRC Practitioner Fellowship. MS Carlino is supported by a Rotary Health Australia scholarship. WA Cooper is supported by Sydney Foundation for Medical Research. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Melanoma Institute Australia, Sydney, New South Wales, Australia M S Carlino, L E Haydu, H Kakavand, A M Menzies, J F Thompson, R F Kefford, R A Scolyer & G V
Long * Westmead Institute for Cancer Research, University of Sydney at Westmead Millennium Institute, Westmead, New South Wales, Australia M S Carlino & R F Kefford * Department of
Medical Oncology, Crown Princess Mary Cancer Centre, Westmead Hospital, Westmead, New South Wales, Australia M S Carlino & R F Kefford * Discipline of Medicine, Sydney Medical School,
The University of Sydney, Sydney, New South Wales, Australia M S Carlino, A M Menzies, A L Hamilton, B Yu, R F Kefford & G V Long * Discipline of Surgery, Sydney Medical School, The
University of Sydney, Sydney, New South Wales, Australia L E Haydu & J F Thompson * Discipline of Pathology, Sydney Medical School, The University of Sydney, Sydney, New South Wales,
Australia H Kakavand, S A O'Toole & R A Scolyer * Department of Medical Oncology, Royal Prince Alfred Hospital, Camperdown, New South Wales, Australia A L Hamilton * Department of
Medical Genomics, Royal Prince Alfred Hospital, Camperdown, New South Wales, Australia B Yu, C C Ng & W A Cooper * Department of Tissue Pathology and Diagnostic Oncology, Royal Prince
Alfred Hospital, Camperdown, New South Wales, Australia W A Cooper, S A O'Toole & R A Scolyer * School of Medicine, University of Western Sydney, Sydney, NSW, Australia W A Cooper *
Department of Melanoma and Surgical Oncology, Royal Prince Alfred Hospital, Camperdown, New South Wales, Australia J F Thompson * The Kinghorn Cancer Centre and Cancer Program Garvan
Institute of Medical Research, Victoria Street, Darlinghurst, New South Wales, Australia S A O'Toole Authors * M S Carlino View author publications You can also search for this author
inPubMed Google Scholar * L E Haydu View author publications You can also search for this author inPubMed Google Scholar * H Kakavand View author publications You can also search for this
author inPubMed Google Scholar * A M Menzies View author publications You can also search for this author inPubMed Google Scholar * A L Hamilton View author publications You can also search
for this author inPubMed Google Scholar * B Yu View author publications You can also search for this author inPubMed Google Scholar * C C Ng View author publications You can also search for
this author inPubMed Google Scholar * W A Cooper View author publications You can also search for this author inPubMed Google Scholar * J F Thompson View author publications You can also
search for this author inPubMed Google Scholar * R F Kefford View author publications You can also search for this author inPubMed Google Scholar * S A O'Toole View author publications
You can also search for this author inPubMed Google Scholar * R A Scolyer View author publications You can also search for this author inPubMed Google Scholar * G V Long View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to M S Carlino. ETHICS DECLARATIONS COMPETING INTERESTS Carlino MS received
honoraria from Novartis, Roche, GlaxoSmithKline and Bristol-Myers Squibb. Menzies AM received travel support from Roche and GlaxoSmithKline, and honoraria from Roche. Hamilton AL received
travel support from Roche. Cooper WA received consultancies and honararia from Pfizer and Merck. O’Toole S received honoraria from Roche, Astra Zeneca and Pfizer. Thompson JF received
consultancies and honoraria from Roche and GlaxoSmithKline. Kefford RF received consultancies and honoraria from Roche, GlaxoSmithKline and Novartis. Scolyer RA received consultancies from
Roche and GlaxoSmithKline, honoraria from Abbott Molecular. Long GV received consultancies from Roche, Bristol-Myers Squibb, GlaxoSmithKline, Novartis and Amgen; honoraria from Roche; travel
support from GlaxoSmithKline and Roche Melanoma Institute Australia received research support from Roche. The remaining authors declare no conflict of interest. ADDITIONAL INFORMATION This
work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons
Attribution-NonCommercial-Share Alike 3.0 Unported License. Supplementary Information accompanies this paper on British Journal of Cancer website SUPPLEMENTARY INFORMATION SUPPLEMENTARY
FIGURE S1 (JPG 148 KB) SUPPLEMENTARY FIGURE S2 (JPG 129 KB) SUPPLEMENTARY FIGURE S3 (JPG 148 KB) SUPPLEMENTARY FIGURE LEGENDS (DOC 42 KB) SUPPLEMENTARY TABLES (DOC 103 KB) RIGHTS AND
PERMISSIONS From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of
this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Carlino, M., Haydu, L., Kakavand, H. _et al._ Correlation
of BRAF and NRAS mutation status with outcome, site of distant metastasis and response to chemotherapy in metastatic melanoma. _Br J Cancer_ 111, 292–299 (2014).
https://doi.org/10.1038/bjc.2014.287 Download citation * Revised: 08 April 2014 * Accepted: 30 April 2014 * Published: 10 June 2014 * Issue Date: 15 July 2014 * DOI:
https://doi.org/10.1038/bjc.2014.287 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * _BRAF_ * _NRAS_ * melanoma * prognosis * BRAF
inhibitors * MEK inhibitors






