- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT BACKGROUND: Immunohistochemistry (IHC) and fluorescent _in situ_ hybridisation (FISH) are currently the most commonly used methods to assess HER2 status. PCR-based assays allow
quantitative determination of HER2 amplification (Q-PCR) or overexpression (Q-RT–PCR), but are not routinely used. We evaluated the relevance of Q-RT–PCR for HER2 status determination.
METHODS: We compared IHC and Q-RT–PCR in 466 breast tumours. In discordant or equivocal cases, five additional methods (IHC with two other antibodies, FISH, silver _in situ_ hybridisation
(SISH) and Q-PCR) were combined to determine HER2 status. Two cases with HER2 intra-tumour heterogeneity were further explored by allelic profiles analysis and HUMARA clonality determination
after microdissection. RESULTS: We observed 97.3% concordance between Q-RT–PCR and non-equivocal IHC. Twelve out of 466 cases (3%) revealed discordances between the two methods. The power
of Q-RT–PCR to predict HER2 status (defined by seven methods) was similar to that of IHC. Although rare, some discordances between techniques might be due to HER2 intra-tumour heterogeneity
and we report two examples, one tumour containing two distinct clones, another tumour consisting of HER2 amplified and non-amplified subclones. CONCLUSION: Q-RT–PCR and IHC are highly
concordant methods for HER2 status assessment, and Q-RT–PCR allows a highly reliable quantitative assessment and could be a useful adjunct to IHC. SIMILAR CONTENT BEING VIEWED BY OTHERS HER2
IN SITU HYBRIDIZATION TEST IN BREAST CANCER: QUANTIFYING MARGINS OF ERROR AND GENETIC HETEROGENEITY Article 12 May 2021 A COMPREHENSIVE APPRAISAL OF HER2 HETEROGENEITY IN _HER2_-AMPLIFIED
AND HER2-LOW COLORECTAL CANCER Article 05 August 2023 _HER2_ AMPLIFICATION AND HER2 LOW EXPRESSION IN ENDOMETRIAL CARCINOMA: PREVALENCE ACROSS MOLECULAR, HISTOLOGICAL AND CLINICOPATHOLOGICAL
RISK GROUPS Article Open access 12 February 2025 MAIN The HER2 gene encodes the human epidermal growth factor receptor 2 with tyrosine kinase activity (Slamon et al, 1987) and is
overexpressed in 15–20% of breast cancers (Owens et al, 2004), through gene amplification with well correlated level of protein expression (Pauletti et al, 1996; Yarden, 2001). This
molecular abnormality defines breast tumours with poor prognosis and increased risk of early relapse (Slamon et al, 1987, 1989), but predicts response to the humanised monoclonal anti-HER2
antibody trastuzumab (Herceptin) or to small tyrosine kinase inhibitors such as lapatinib or erlotinib. The increasing number of patients with breast cancer whose survival has been improved
by trastuzumab treatment underlines the need for sensitive, specific, highly reproducible and cost-efficient methods to identify patients eligible for anti-HER2 therapies. Furthermore, HER2
status not only predicts anti-HER2 efficacy, but could also determine other treatment options. Indeed, HER2-overexpressing breast tumours are often resistant to hormonotherapy and more
sensitive to anthracycline-based and taxane-containing chemotherapy (Paik et al, 2000; Pritchard et al, 2008), so that all invasive breast cancer need an HER2 evaluation at diagnosis. In
daily practice, according to the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAPs) recommandations (Wolff et al, 2007), HER2 status is determined by
immunohistochemistry (IHC) followed, if necessary, by fluorescent _in situ_ hybridisation (FISH), although FISH first-line determination is also encouraged by some authors (Sauter et al,
2009). Immunohistochemistry results are obtained on formalin-fixed paraffin-embedded (FFPE) samples and expressed as a four-scale score system (0 to 3+). Determination of HER2 status by FISH
is also performed on FFPE samples. It shows the mean number of HER2 copies using a DNA probe hybridising to the HER2 gene alone or in association with a centromeric probe as control for
chromosome 17 copy number expressed as HER2/CEN17 ratio. Fluorescent _in situ_ hybridisation is now challenged by chromogenic (CISH) or silver _in situ_ hybridisation (SISH), faster methods
using a chromogenic signal that do not decay over time, that can be further reevaluated and need only a classical light microscope (Isola et al, 2004; Laakso et al, 2006; Papouchado et al,
2010). A ‘gold standard’ for HER2 determination does not really exist and the ASCO/CAPs study estimates that 20% of current HER2 tests may be inaccurate owing to multiple preanalytic and
analytic variables (Wolff et al, 2007). Indeed, the existence of various IHC protocols, FDA-approved antibodies or probes contributes to interlaboratory variability. Improvement in
HER2-testing reproducibility between laboratories is crucial and new HER2-testing technologies are also needed. Different studies have evaluated the performance of Q-RT–PCR or Q-PCR to
determine the HER2 status (Laudadio et al, 2007). Quantitative PCR using primers flanking the HER2 gene or mRNA can quantify gene amplification or messenger overexpression. PCR-based assays
are easy, rapid, sensitive, specific and quantitative approaches without the inherent inter-observer variability of IHC and FISH/CISH techniques. It can be used on small samples and can be
standardised and automated. DNA extraction can also be performed on FFPE tissues. However, RT–PCR technology, more sensitive to RNA quality, is more robust if frozen tissues are used. The
main PCR drawback is that DNA or RNA extraction mixes tumour and non-tumour cells and can lead to tumour cells dilution with risk of false negative. Conversely, the mix of invasive tumour
cells with high-grade intraductal HER2-positive tumour cells can lead to false positive results. Provided that sample purity is controlled with microscopical examination, PCR-based
determination has been shown to correlate well with IHC and FISH (Vinatzer et al, 2005; Barberis et al, 2008). In order to evaluate the benefits of PCR-based technology in daily HER2
testing, we performed a large prospective study comparing HER2 determination with IHC and Q-RT–PCR. As recommended in guidelines, IHC 2+ cases were further explored not only by FISH and SISH
but also by other IHC tests and Q-PCR. Moreover, all discordant cases were explored by these additional techniques. We also analysed extensively two cases showing striking HER2 intra-tumour
heterogenity that may explain some technical discordances. We observed that molecular approaches are powerful and reliable quantitative tools for HER2 status assessment that could
complement IHC for optimal patient care. MATERIALS AND METHODS PATIENTS AND SAMPLES We analysed 466 primary breast tumours obtained from patients treated in Saint Louis Hospital (Paris) from
2002 to 2007. All patients were informed of the study according to our Institutional Review Board recommendations. A total of 332 samples were obtained from surgical specimen and 134 were
obtained with fine-needle biopsies. Tumours with >10% _in situ_ component were excluded from this study. Haematoxylin–eosin (H&E) stainings, immunohistochemical stainings and _in
situ_ hybridisation techniques were performed on FFPE tissue samples. Q-RT–PCR and Q-PCR were performed on RNA and DNA extracted from frozen tissues. IHC DETECTION HER2 immunohistochemistry
was performed with the monoclonal HER2 CB11 antibody (Novocastra, Newcastle upon Tyne, UK, dilution 1/250) in the BenchmarkXT immunostainer (Roche Diagnostics, Basel, Switzerland) with
calibrated positive controls and internal (on slide) negative controls. Evaluation of immunostainings was performed by two pathologists (PB, AR) and scored according to ASCO guidelines
(Wolff et al, 2007; Gown, 2008): negative for 0 (no membrane staining) and 1+ (faint or barely perceptible incomplete membrane staining); equivocal for 2+ (10–30% tumour cells with strong
complete membrane staining or >10% tumour cells with moderate complete membrane staining) and positive for 3+ (>30% tumour cells with strong complete membrane staining). For discordant
and CB11 equivocal cases, HER2 immunohistochemistry was performed with A0485 polyclonal (Dako, Glostrup, Denmark, dilution 1/500) and 4B5 monoclonal antibody (Roche Diagnostics, prediluted)
using the Discovery immunostainer (Roche Diagnostics). HER2 scores were evaluated as described below. Other antibodies were used with the Discovery immunostainer: oestrogen receptor
(Novocastra, clone 6F11, dilution 1/50), progesteron receptor (Novocastra, clone 312, dilution 1/75), cytokeratin 5/6 (Dako, clone B4, dilution 1/50), cytokeratin 17 (Dako, clone E30,
dilution 1/50) and cytokeratin 8 (Millipore, Billerica, MA, USA, clone MAB 3414, 1/50). SISH AND FISH DETECTION Silver _in situ_ hybridisation and FISH were performed on 3 _μ_m paraffin
tissue sections. Silver _in situ_ hybridisation staining, with HER2 and chromosome 17 probes, was performed in BenchmarkXT slide stainers (Roche Diagnostics) and described in Dietel et al
(2007). Fluorescent _in situ_ hybridisation staining was performed using the Zytolight Spec HER2/CEN17 kit (Zytovision, CliniScience, Montrouge, France) according to the manufacturer's
protocol. Fluorescence signal were counted by one pathologist (MA) using a (Leica DM 4000) Zeiss Axioscope Imager Z1 fluorescence microscope (Zeiss, Oberkochen, Germany). A minimum of 80
tumour cell nuclei, with intact morphology according to DAPI counterstaining, were counted. The HER2/CEN17 ratio was obtained by dividing the mean number of HER2 signals by the mean number
of CEN17 signals in tumour cells and defined HER2 gene amplification if >2.2, equivocal if between 1.8 and 2.2 and no HER2 amplification if <1.8, according to ASCO/CAPs recommendations
(Wolff et al, 2007). QUANTIFICATION OF HER2 OVEREXPRESSION BY Q-RT–PCR AND HER2 GENE COPY NUMBER BY Q-PCR Nucleic acids were extracted by phenol/chloroform procedure. Tumour cell purity and
presence of _in situ_ carcinoma were assessed on adjacent H&E-stained sections. Quantitative PCR were performed on LightCycler 2.1 instrument (Roche Diagnostics). HER2 overexpression
was evaluated by relative quantification using TATA-binding protein as endogen control (Bossard et al, 2005). Final result was expressed as a normalised ratio considered as over-expressed if
>7. The cut-off ratio was determined on a tumours training set using univariate partition method (XLSTAT software) and correlation with IHC–HER2 expression. HER2 amplification was
evaluated on DNA using the LightCycler-HER-2/neu DNA Quantification kit (Roche Diagnostics) in all IHC2+ and IHC/Q-RT–PCR discordant specimens. The assay amplifies simultaneously one HER2
fragment and one gastrin fragment, the reference gene localised on the chromosome 17. Results were expressed as the ratio of HER2/gastrin in the sample, normalised with the same ratio in the
calibrator DNA set. A ratio above 2 was considered amplified. Results between 2 and 3 were repeated. ALLELIC PROFILES ANALYSIS Allelic profiles were analysed as described in Varna et al
(2007). We used a PALM Microbeam/Olympus system to perform laser tissue microdissection on FFPE tissue sections. PCR was performed directly on cell lysates with at least 500 cells for each
PCR. Five microsatellite dinucleotide repeats were used: D17S250, D17S855, D17S1840, D13S153 and D9S171. Whole tumour allelic profiles and microdissected areas allelic profiles were
compared. CLONALITY ASSESSMENT USING ANDROGEN RECEPTOR GENE METHYLATION PATTERN The androgen receptor gene (HUMARA) polymorphism is characterised by highly polymorphic short-tandem CAG
repeat units, 100 bp downstream of a methylated site in the coding region of its first exon (Lucas et al, 1997; Fujita et al, 1998; Wang et al, 2007). Before digesting the genomic DNA with
methylation-sensitive restriction enzyme HpaII, electrophoresis of heterozygote cases shows two alleles. After digesting the genomic DNA with HpaII, electrophoresis shows two different
alleles if the tumour is not monoclonal and only one allele or at least strong allelic imbalance if the tumour is of monoclonal origin due to X-chromosome inactivation mosaicism, as solely
the inactive methylated allele is not cut at the restriction site and hence PCR amplified. Tumour DNA was amplified at the HUMARA locus either with or without HpaII predigestion and overall
tumour profiles and microdissected areas profiles were compared. RESULTS IHC AND Q-RT–PCR COMPARISONS To determine HER2 status, we performed immunohistochemistry with CB11 antibody and
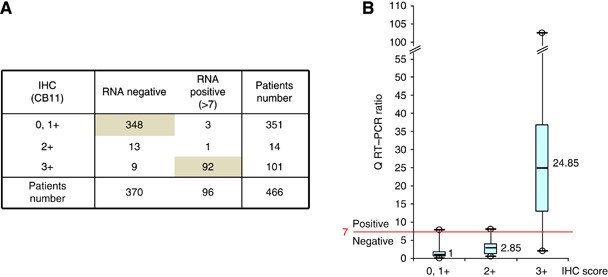
Q-RT–PCR on all 466 cases (Figure 1). Overall concordance was excellent (97.3%), especially in IHC 0,1+ subgroup (348 negative Q-RT–PCR/351 IHC 0/1+: 99.2%). Concordance was also good in IHC
3+ cases (91%: 92 positive Q-RT–PCR/101 IHC 3+). However, in 12 out of 466 cases (3%), the two techniques were discordant (either IHC 0,1+/Q-RT–PCR>7 or IHC 3+/Q-RT–PCR<7). Among
these 12 discordant cases, 11 had been obtained after surgical procedure and only one after fine-needle biopsy. In the 14 out of 466 IHC 2+ equivocal cases, Q-RT–PCR showed the absence of
HER2 overexpression in 13 out of 14 cases (93%). ANALYSIS OF THE 26 DISCORDANT OR EQUIVOCAL CASES According to guidelines, all IHC score 2+ (_n_=14) were analysed not only by hybridisation
methods (FISH and SISH), but also with two other IHC–HER2 antibodies and Q-PCR (Figure 2). These additional methods were also performed in all discordant cases (_n_=12). An overall HER2
status was defined for each of these 26 discordant or equivocal cases, based on the results of all seven techniques used, a case being HER2 positive if there were more positive than negative
results (12 out of 26 cases), being negative if there were more negative than positive results (12 out of 26 cases) and being HER2 unclassified in other situations (2 out of 26 cases).
Among the 14 equivocal cases, 4 were finally scored positive by the overall HER2 status, 9 negative and 1 remained undefined. In these equivocal cases, Q-RT–PCR analysis predicted the final
HER2 status in 10 cases and failed in only 3 cases. However, in the 12 discordant cases, Q-RT–PCR predicted final HER2 status in only two cases and failed in nine cases. The overall ability
of Q-RT–PCR to predict final HER2 status in the 24 cases with known final HER2 status was, therefore, 12 out of 24, while the overall ability of IHC CB11 to predict final HER2 status was 9
out of 24. Accordingly, Q-RT–PCR and IHC, respectively, predicted the overall HER2 status in 452 tumours and in 449 tumours. Among the 26 IHC/Q-RT–PCR discordant or IHC 2+ cases, only six
tumours (#1, 2, 14, 23, 24 and 25) showed highly concordant results, with 6 out of 7 methods showing similar results. The other 20 cases all showed more extensive discrepancies among the
techniques used, with only three to five concordant methods. The results in case #4, either positive or borderline, were possibly due to chromosome 17 polysomy that was demonstrated with
FISH and SISH techniques. In case #17 (Figure 2B), we observed heterogeneous IHC staining among the three antibodies used, as well as FISH negativity and moderate amplification with SISH. In
the other cases, no easy explanation could be given for these technical discrepancies and either borderline HER2 status or true intra-tumour heterogeneity could be implicated. PHENOTYPE AND
GENOTYPE ANALYSES OF TWO CASES WITH HER2 INTRA-TUMOUR HETEROGENEITY In order to explore one possible cause of discordances between techniques, we analysed two cases that showed obvious
intra-tumour HER2 heterogeneity. In _Case A_, H&E examination showed the presence of a 1-cm less differentiated area (area 1) located inside the main tumour component (area 2) (Figure
3A). Area 1 was scored 3+ for HER2, was positive for CK5/6 and negative for ER, PR, CK8 and CK17, while area 2 was negative for HER2, CK5/6 and CK17 and positive for ER, PR and CK8. Silver
_in situ_ hybridisation confirmed HER2 amplification only in tumour cells of area 1 (Figure 3A). There was allelic loss at D17S855 in area 1 but not in area 2 (data not shown). Allelic
profiles with the other microsatellites were either non-informative or showed no significant difference between the two areas (data not shown). The analysis of the X-chromosome methylation
pattern showed important differences between areas 1 and 2 (Figure 3B): before HpaII digestion, allelic profiles showed LOH in each area, but on two distinct alleles. After HpaII digestion,
profiles showed inactivation of one X chromosome in both areas 1 and 2, indicating that both areas are populated by monoclonal cells. However, areas 1 and 2 did not inactivate the same X
chromosome, demonstrating that they are not deriving from the same clone. In _Case B_ (case #18 of this study), HER2 immunohistochemical staining (Figure 4A) showed two sharply demarcated
areas, one with a strong membranous staining in over 80% of tumour cells (area 1) and one totally negative (area 2). These two areas looked totally similar on H&E staining. Other
immunostainings (ER, PR, CK5/6, CK17 and CK8) were similar in both areas (data not shown). Silver _in situ_ hybridisation confirmed HER2 amplification in area 1 and lack of amplification in
area 2, with sharp borders between the two areas (Figure 4A). Allelic profiles obtained after microdissection with D17S1840 showed only one allele in area 1 and only the other allele in area
2, consistent with subclonal heterogeneity (Figure 4B). For D17S250, one new allele was observed in the non-microdissected tumour, as well as in areas 1 and 2 (Figure 4B), strongly
suggesting that areas 1 and 2 are derived from a common tumour cell. Allelic profiles with the three other microsatellites were almost similar in both areas. The analysis of the X-chromosome
methylation pattern was not informative since it showed inactivation of the same X chromosome in both areas (data not shown). DISCUSSION Selection of patients for trastuzumab treatment is
primarily performed by IHC using HercepTest, CB11 or 4B5 antibody, as recommended (Birner et al, 2001; Wolff et al, 2007). HER2 immunostaining is easy to perform, available as a standard
method in pathology laboratories, widely applicable (on FFPE specimens), very reliable (Lebeau et al, 2010; Purdie et al, 2010) and relatively inexpensive, but is only a semi-quantitative
method. Yet, in 3–15% of cases (Dendukuri et al, 2007), IHC is equivocal and further analyses are required, leading to the usual IHC+FISH association. Like IHC, FISH is a semi-quantitative
morphological method but with higher costs and need of specialised expertise and equipment. In the context of HER2 status assessment, Q-RT–PCR could also be a useful option and an
alternative to the current IHC+FISH procedure. Several studies have already compared IHC and Q-RT–PCR (Cronin et al, 2004; Ginestier et al, 2004; Gjerdrum et al, 2004; Bossard et al, 2005;
Esteva et al, 2005; Vanden Bempt et al, 2005; Vinatzer et al, 2005; Bergqvist et al, 2007; Kostopoulou et al, 2007; Barberis et al, 2008; Cuadros et al, 2010) (Table 1). These reports showed
good overall concordance (82–100%) with frozen or FFPE specimen, and mostly without microdissection, but in small patient series. Moreover, HER2 mRNA evaluation was shown to be a fast,
reliable and cost-effective alternative to the IHC+FISH procedure and also correlated with overall survival and disease-free survival (Vinatzer et al, 2005). In this prospective study of 466
breast tumours comparing IHC and Q-RT–PCR determination of HER2 status, we show that Q-RT–PCR was very strongly correlated with IHC (overall concordance 97.3%). In the 12 discordant cases,
Q-RT–PCR was not as powerful as IHC to predict the final HER2 status determined by five other methods, with seven and one false negative cases and two and one false positive cases for
Q-RT–PCR and IHC, respectively. However, four cases (Figure 2A, cases #23, 24, 25 and 26) among the seven Q-RT–PCR false negative cases had ratios very close to the cut-off value, suggesting
that tumour cells dilution could explain the discrepancy. Note that in cases #23, 24 and 25, only Q-RT–PCR method was unable to predict the HER2 positivity, perhaps because mRNA analysis
can be tricky in borderline cases. Similarly, in the two false positive cases (#1 and #2), the ratio was also close over the cut-off value and #1 presented a small _in situ_ component,
although evaluated to be <10%. More importantly, in the 14 IHC equivocal cases, Q-RT–PCR was highly predictive of the final HER2 status in 10 out of 13 cases. These results thus validate
the use of Q-RT–PCR as alternative to FISH in IHC 2+ cases. Overall, Q-RT–PCR and IHC had statistically similar efficiency for predicting HER2 status in the overall 466 cases we studied (452
out of 466 and 449 out of 466, respectively). Q-RT–PCR is quick, easy to perform, quantitative and has no inter-observer variability. However, dilution of tumour genomic material with
non-neoplastic tissue or presence of _in situ_ component are well-known drawbacks of Q-RT–PCR, so that microscopical control of the sample is particularly crucial prior to molecular
extraction. If expert pathological selection is performed, microdissection can clearly be avoided (Gjerdrum et al, 2004). Since Q-RT–PCR can be performed with as little as 100 ng RNA, the
corresponding amount of frozen tissue is easily obtained even with 14G or 16G fine-needle biopsies of breast tumours (O'Flynn et al, 2010), although a frozen tissue workflow has to be
organised. While we determined here the mRNA level using frozen tissues, several reports demonstrated also the good HER2 mRNA/protein concordance in FFPE samples (Cronin et al, 2004;
Barberis et al, 2008). Immunohistochemistry and Q-RT–PCR are, therefore, two complementary approaches, with an excellent overall sensitivity and with almost no equivocal cases. Measurement
of tumour cell percentage and morphological HER2 assessment are done by H&E and IHC stainings, while quantitative HER2 assessment is obtained by Q-RT–PCR, for moderate costs (Vinatzer et
al, 2005). Indeed, the combination of IHC+Q-RT–PCR has an estimated cost of 157.27 euros, compared with 525.68 euros for IHC+FISH determination (Vinatzer et al, 2005). The association of
these two techniques in breast cancer would be useful for patient care, each technique controlling and complementing the other one. However, it is not possible to draw similar conclusions in
gastroesophageal cancers, now often tested for HER2 status, since these tumours might be more heterogeneous than breast tumours and the use of molecular techniques in this field of
pathology should be evaluated. One rare but significant finding in our study was that a few tumours (12 out of 466) had a highly discordant HER2 status depending on the methods used. One of
these discordant cases (tumour #4) showed chromosome 17 polysomy that is well known to complicate HER2 status analysis (Dal Lago et al, 2006). Another case (tumour #17) showed highly
discordant results among the three antibodies used for IHC, although stainings had been performed on three adjacent tissue sections, maybe reflecting inappropriate fixation of the sample.
For the other 10 discordant cases, only hypotheses can be drawn to explain these discrepancies: some of these cases could have a true borderline HER2 status; therefore, being considered
negative or positive with only slight technical sensitivity changes. Intra-tumour heterogeneity may also explain some of these discrepancies, even when the analyses are performed on close
tissue areas. We describe here two cases that are typical examples of regional HER2 intra-tumour heterogeneity. In case A, methylation of one X chromosome in HER2+ area and methylation of
the other X chromosome in HER2− area strongly suggest the existence of two distinct tumours, deriving from two distinct initiating tumour cells. Case A would be, therefore, like the
so-called ‘collision tumours’ described by pathologists (Isaka et al, 2007; Kleist et al, 2010) all characterised by the simultaneous occurrence at the same place and at the same time of two
tumours with distinct histological types. In case B, the presence in both HER2+ and HER2− areas of shared new allelic abnormalities strongly suggests that the tumour derives from a single
clone, HER2+ area and HER2− area being two different subclones. Therefore, in case B, HER2 amplification may be a progression event in a clonal tumour cell population, as recently reported
(Hanna et al, 2007; Kostopoulou et al, 2007; Cottu et al, 2008; Apple et al, 2009). In conclusion, we demonstrate an excellent concordance between IHC and Q-RT–PCR for HER2 status assessment
in breast tumours. Rare discordances may be due sometimes to intra-tumour heterogeneity. The association of these two methods in IHC equivocal cases or even in all tumours may be a reliable
and moderate-cost strategy for HER2 status assessment. The quantitative nature of Q-RT–PCR could also provide clinically relevant informations, allowing tailored treatment according to the
amplitude of HER2 overexpression in breast cancers. CHANGE HISTORY * _ 29 MARCH 2012 This paper was modified 12 months after initial publication to switch to Creative Commons licence terms,
as noted at publication _ REFERENCES * Apple SK, Lowe AC, Rao PN, Shintaku IP, Moatamed NA (2009) Comparison of fluorescent _in situ_ hybridization HER-2/neu results on core needle biopsy
and excisional biopsy in primary breast cancer. _Mod Pathol_ 22: 1151–1159 Article CAS PubMed Google Scholar * Barberis M, Pellegrini C, Cannone M, Arizzi C, Coggi G, Bosari S (2008)
Quantitative PCR and HER2 testing in breast cancer: a technical and cost-effectiveness analysis. _Am J Clin Pathol_ 129: 563–570 Article PubMed Google Scholar * Bergqvist J, Ohd JF, Smeds
J, Klaar S, Isola J, Nordgren H, Elmberger GP, Hellborg H, Bjohle J, Borg AL, Skoog L, Bergh J (2007) Quantitative real-time PCR analysis and microarray-based RNA expression of HER2 in
relation to outcome. _Ann Oncol_ 18: 845–850 Article CAS PubMed Google Scholar * Birner P, Oberhuber G, Stani J, Reithofer C, Samonigg H, Hausmaninger H, Kubista E, Kwasny W,
Kandioler-Eckersberger D, Gnant M, Jakesz R (2001) Evaluation of the United States Food and Drug Administration-approved scoring and test system of HER-2 protein expression in breast cancer.
_Clin Cancer Res_ 7: 1669–1675 CAS PubMed Google Scholar * Bossard C, Bieche I, Le Doussal V, Lidereau R, Sabourin JC (2005) Real-time RT-PCR: a complementary method to detect HER-2
status in breast carcinoma. _Anticancer Res_ 25: 4679–4683 CAS PubMed Google Scholar * Cottu PH, Asselah J, Lae M, Pierga JY, Dieras V, Mignot L, Sigal-Zafrani B, Vincent-Salomon A (2008)
Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. _Ann Oncol_ 19: 595–597 Article CAS PubMed Google Scholar * Cronin
M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB (2004) Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene
reverse transcriptase-polymerase chain reaction assay. _Am J Pathol_ 164: 35–42 Article CAS PubMed PubMed Central Google Scholar * Cuadros M, Talavera P, Lopez FJ, Garcia-Perez I,
Blanco A, Concha A (2010) Real-time RT-PCR analysis for evaluating the Her2/neu status in breast cancer. _Pathobiology_ 77: 38–45 Article CAS PubMed Google Scholar * Dal Lago L, Durbecq
V, Desmedt C, Salgado R, Verjat T, Lespagnard L, Ma Y, Veys I, Di Leo A, Sotiriou C, Piccart M, Larsimont D (2006) Correction for chromosome-17 is critical for the determination of true
Her-2/neu gene amplification status in breast cancer. _Mol Cancer Ther_ 5: 2572–2579 Article CAS PubMed Google Scholar * Dendukuri N, Khetani K, McIsaac M, Brophy J (2007) Testing for
HER2-positive breast cancer: a systematic review and cost-effectiveness analysis. _CMAJ_ 176: 1429–1434 Article PubMed PubMed Central Google Scholar * Dietel M, Ellis IO, Hofler H,
Kreipe H, Moch H, Dankof A, Kolble K, Kristiansen G (2007) Comparison of automated silver enhanced _in situ_ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene
status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. _Virchows Arch_ 451: 19–25 Article CAS PubMed
Google Scholar * Esteva FJ, Sahin AA, Cristofanilli M, Coombes K, Lee SJ, Baker J, Cronin M, Walker M, Watson D, Shak S, Hortobagyi GN (2005) Prognostic role of a multigene reverse
transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. _Clin Cancer Res_ 11: 3315–3319 Article CAS PubMed Google Scholar * Fujita
MQ, Hashida N, Shin M, Nakanishi H, Yoshihara W, Aozasa K (1998) Clonal composition of malignant fibrous histiocytoma: analysis by PCR-based assay of the human androgen receptor gene
(HUMARA). _Oncology_ 55: 600–606 Article CAS PubMed Google Scholar * Ginestier C, Charafe-Jauffret E, Penault-Llorca F, Geneix J, Adelaide J, Chaffanet M, Mozziconacci MJ, Hassoun J,
Viens P, Birnbaum D, Jacquemier J (2004) Comparative multi-methodological measurement of ERBB2 status in breast cancer. _J Pathol_ 202: 286–298 Article CAS PubMed Google Scholar *
Gjerdrum LM, Sorensen BS, Kjeldsen E, Sorensen FB, Nexo E, Hamilton-Dutoit S (2004) Real-time quantitative PCR of microdissected paraffin-embedded breast carcinoma: an alternative method for
HER-2/neu analysis. _J Mol Diagn_ 6: 42–51 Article CAS PubMed PubMed Central Google Scholar * Gown AM (2008) Current issues in ER and HER2 testing by IHC in breast cancer. _Mod Pathol_
21 (Suppl 2): S8–S15 Article CAS PubMed Google Scholar * Hanna W, Nofech-Mozes S, Kahn HJ (2007) Intratumoral heterogeneity of HER2/neu in breast cancer – a rare event. _Breast J_ 13:
122–129 Article CAS PubMed Google Scholar * Isaka T, Nakamura T, Tajika M, Kawai H, Imaoka H, Okamoto Y, Aoki M, Inoue H, Takahashi K, Mizuno N, Sawaki A, Yamao K, Seto M, Yokoi T,
Yatabe Y, Nakamura S (2007) API2-MALT1 chimeric transcript-positive gastroduodenal MALT lymphoma with subsequent development of adenocarcinoma as a collision tumour over a clinical course of
7 years. _Histopathology_ 51: 119–123 Article CAS PubMed Google Scholar * Isola J, Tanner M, Forsyth A, Cooke TG, Watters AD, Bartlett JM (2004) Interlaboratory comparison of HER-2
oncogene amplification as detected by chromogenic and fluorescence _in situ_ hybridization. _Clin Cancer Res_ 10: 4793–4798 Article CAS PubMed Google Scholar * Kleist B, Lasota J,
Miettinen M (2010) Gastrointestinal stromal tumor and gastric adenocarcinoma collision tumors. _Hum Pathol_ 41: 1034–1039 Article PubMed Google Scholar * Kostopoulou E, Vageli D,
Kaisaridou D, Nakou M, Netsika M, Vladica N, Daponte A, Koukoulis G (2007) Comparative evaluation of non-informative HER-2 immunoreactions (2+) in breast carcinomas with FISH, CISH and
QRT-PCR. _Breast_ 16: 615–624 Article PubMed Google Scholar * Laakso M, Tanner M, Isola J (2006) Dual-colour chromogenic _in situ_ hybridization for testing of HER-2 oncogene
amplification in archival breast tumours. _J Pathol_ 210: 3–9 Article CAS PubMed Google Scholar * Laudadio J, Quigley DI, Tubbs R, Wolff DJ (2007) HER2 testing: a review of detection
methodologies and their clinical performance. _Expert Rev Mol Diagn_ 7: 53–64 Article CAS PubMed Google Scholar * Lebeau A, Turzynski A, Braun S, Behrhof W, Fleige B, Schmitt WD, Grob
TJ, Burkhardt L, Holzel D, Jackisch C, Thomssen C, Muller V, Untch M (2010) Reliability of human epidermal growth factor receptor 2 immunohistochemistry in breast core needle biopsies. _J
Clin Oncol_ 28: 3264–3270 Article PubMed Google Scholar * Lucas DR, Shroyer KR, McCarthy PJ, Markham NE, Fujita M, Enomoto TE (1997) Desmoid tumor is a clonal cellular proliferation: PCR
amplification of HUMARA for analysis of patterns of X-chromosome inactivation. _Am J Surg Pathol_ 21: 306–311 Article CAS PubMed Google Scholar * O'Flynn EA, Wilson AR, Michell MJ
(2010) Image-guided breast biopsy: state-of-the-art. _Clin Radiol_ 65: 259–270 Article CAS PubMed Google Scholar * Owens MA, Horten BC, Da Silva MM (2004) HER2 amplification ratios by
fluorescence _in situ_ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. _Clin Breast Cancer_ 5: 63–69 Article CAS PubMed Google Scholar
* Paik S, Bryant J, Tan-Chiu E, Yothers G, Park C, Wickerham DL, Wolmark N (2000) HER2 and choice of adjuvant chemotherapy for invasive breast cancer: National Surgical Adjuvant Breast and
Bowel Project Protocol B-15. _J Natl Cancer Inst_ 92: 1991–1998 Article CAS PubMed Google Scholar * Papouchado BG, Myles J, Lloyd RV, Stoler M, Oliveira AM, Downs-Kelly E, Morey A,
Bilous M, Nagle R, Prescott N, Wang L, Dragovich L, McElhinny A, Garcia CF, Ranger-Moore J, Free H, Powell W, Loftus M, Pettay J, Gaire F, Roberts C, Dietel M, Roche P, Grogan T, Tubbs R
(2010) Silver _in situ_ hybridization (SISH) for determination of HER2 gene status in breast carcinoma: comparison with FISH and assessment of interobserver reproducibility. _Am J Surg
Pathol_ 34: 767–776 Article PubMed Google Scholar * Pauletti G, Godolphin W, Press MF, Slamon DJ (1996) Detection and quantitation of HER-2/neu gene amplification in human breast cancer
archival material using fluorescence _in situ_ hybridization. _Oncogene_ 13: 63–72 CAS PubMed Google Scholar * Pritchard KI, Messersmith H, Elavathil L, Trudeau M, O'Malley F,
Dhesy-Thind B (2008) HER-2 and topoisomerase II as predictors of response to chemotherapy. _J Clin Oncol_ 26: 736–744 Article CAS PubMed Google Scholar * Purdie CA, Jordan LB, McCullough
JB, Edwards SL, Cunningham J, Walsh M, Grant A, Pratt N, Thompson AM (2010) HER2 assessment on core biopsy specimens using monoclonal antibody CB11 accurately determines HER2 status in
breast carcinoma. _Histopathology_ 56: 702–707 Article PubMed Google Scholar * Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF (2009) Guidelines for human epidermal growth factor
receptor 2 testing: biologic and methodologic considerations. _J Clin Oncol_ 27: 1323–1333 Article CAS PubMed Google Scholar * Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire
WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. _Science_ 235: 177–182 Article CAS PubMed Google Scholar * Slamon DJ,
Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer.
_Science_ 244: 707–712 Article CAS PubMed Google Scholar * Vanden Bempt I, Vanhentenrijk V, Drijkoningen M, Wlodarska I, Vandenberghe P, De Wolf-Peeters C (2005) Real-time reverse
transcription-PCR and fluorescence _in-situ_ hybridization are complementary to understand the mechanisms involved in HER-2/neu overexpression in human breast carcinomas. _Histopathology_
46: 431–441 Article CAS PubMed Google Scholar * Varna M, Soliman H, Feugeas JP, Turpin E, Chapelin D, Legres L, Plassa LF, de Roquancourt A, Espie M, Misset JL, Janin A, de The H,
Bertheau P (2007) Changes in allelic imbalances in locally advanced breast cancers after chemotherapy. _Br J Cancer_ 97: 1157–1164 Article CAS PubMed PubMed Central Google Scholar *
Vinatzer U, Dampier B, Streubel B, Pacher M, Seewald MJ, Stratowa C, Kaserer K, Schreiber M (2005) Expression of HER2 and the coamplified genes GRB7 and MLN64 in human breast cancer:
quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence _in situ_ hybridization. _Clin Cancer Res_ 11: 8348–8357 Article CAS
PubMed Google Scholar * Wang EH, Dai SD, Qi FJ, Hong-Tao X, Wei Q (2007) Gene expression and clonality analysis of the androgen receptor and phosphoglycerate kinase genes in polygonal
cells and cuboidal cells in so-called pulmonary sclerosing hemangioma. _Mod Pathol_ 20: 1208–1215 Article CAS PubMed Google Scholar * Wolff AC, Hammond ME, Schwartz JN, Hagerty KL,
Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M,
Wheeler TM, Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists Guideline recommendations for human epidermal growth factor receptor 2 testing in breast
cancer. _Arch Pathol Lab Med_ 131: 18 CAS PubMed Google Scholar * Yarden Y (2001) Biology of HER2 and its importance in breast cancer. _Oncology_ 61 (Suppl 2): 1–13 Article CAS PubMed
Google Scholar Download references ACKNOWLEDGEMENTS We thank all the technicians of St Louis hospital Pathology Department and the technicians of the Biochemistry Department (Catherine
Brunin, Dominique Chapelin, Odile Flinois, Laurence Françoise, Brice Geslot, Martine Legrand and Evelyne Wittmer). We also thank Jean Paul Feugeas for statistical advices. This work is
dedicated to our friend and colleague Elisabeth Turpin, who initiated this work and who passed away in March 2010 after a long and painful battle against disease. This work was supported by
Programme Hospitalier de Recherche Clinique (PHRC), the Association pour la Recherche sur le Cancer (ARC), the Institut National du Cancer (INCa) and the Région Ile de France. AUTHOR
CONTRIBUTIONSJL-C and PB have designed the study, performed experiments, analysed the results and written the manuscript. FA-B has performed immunohistochemistry, SISH and microdissection.
ET has designed the study and performed Q-RT–PCR experiments. MA and EF have performed and analysed FISH experiments. HS has performed and analysed microsatellite and HUMARA techniques. LL
and MV have performed tissue microdissection. CB has performed Q-RT–PCR experiments. RB has performed immunohistochemical and SISH techniques. AR has analysed IHC experiments and read the
manuscript. L-FP has analysed Q-RT–PCR results and read the manuscript. SG has collected patients data, analysed results and read the manuscript. ME, CB, LC-D and EB have collected patients
data and read the manuscript. AJ has read the manuscript. HT has designed the study, analysed the results and wrote the manuscript. AUTHOR INFORMATION Author notes * E Turpin: Deceased.
AUTHORS AND AFFILIATIONS * Department of Biochemistry, AP-HP, Hosp Saint-Louis, Paris, 75010, France J Lehmann-Che, E Turpin, H Soliman, C Bocquet, L-F Plassa & H de Thé * CNRS
UMR7212/INSERMU944, Paris, 75010, France J Lehmann-Che, E Turpin & H de Thé * Univ Paris Diderot, Sorbonne Paris Cité, Paris, 75010, France J Lehmann-Che, E Turpin, A de Roquancourt, A
Janin, H de Thé & P Bertheau * Department of Pathology, AP-HP, Hosp Saint-Louis, Paris, 75010, France F Amira-Bouhidel, A de Roquancourt, A Janin & P Bertheau * INSERM UMR_S728,
Paris, 75010, France F Amira-Bouhidel, L Legres, R Bernoud, M Varna, A de Roquancourt, A Janin & P Bertheau * Department of Pathology, AP-HP, Hosp Tenon, Paris, 75010, France M Antoine
& E Flandre * AP-HP, Hosp Saint-Louis, Breast Diseases Center, Paris, 75010, France S Giacchetti, M Espié & E Bourstyn * AP-HP, Hosp Saint-Louis, Department of Radiology, Paris,
75010, France C de Bazelaire * AP-HP, Hosp Saint-Louis, Department of Surgery, Paris, 75010, France L Cahen-Doidy Authors * J Lehmann-Che View author publications You can also search for
this author inPubMed Google Scholar * F Amira-Bouhidel View author publications You can also search for this author inPubMed Google Scholar * E Turpin View author publications You can also
search for this author inPubMed Google Scholar * M Antoine View author publications You can also search for this author inPubMed Google Scholar * H Soliman View author publications You can
also search for this author inPubMed Google Scholar * L Legres View author publications You can also search for this author inPubMed Google Scholar * C Bocquet View author publications You
can also search for this author inPubMed Google Scholar * R Bernoud View author publications You can also search for this author inPubMed Google Scholar * E Flandre View author publications
You can also search for this author inPubMed Google Scholar * M Varna View author publications You can also search for this author inPubMed Google Scholar * A de Roquancourt View author
publications You can also search for this author inPubMed Google Scholar * L-F Plassa View author publications You can also search for this author inPubMed Google Scholar * S Giacchetti View
author publications You can also search for this author inPubMed Google Scholar * M Espié View author publications You can also search for this author inPubMed Google Scholar * C de
Bazelaire View author publications You can also search for this author inPubMed Google Scholar * L Cahen-Doidy View author publications You can also search for this author inPubMed Google
Scholar * E Bourstyn View author publications You can also search for this author inPubMed Google Scholar * A Janin View author publications You can also search for this author inPubMed
Google Scholar * H de Thé View author publications You can also search for this author inPubMed Google Scholar * P Bertheau View author publications You can also search for this author
inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to J Lehmann-Che. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. RIGHTS AND PERMISSIONS From
twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license,
visit http://creativecommons.org/licenses/by-nc-sa/3.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lehmann-Che, J., Amira-Bouhidel, F., Turpin, E. _et al._
Immunohistochemical and molecular analyses of HER2 status in breast cancers are highly concordant and complementary approaches. _Br J Cancer_ 104, 1739–1746 (2011).
https://doi.org/10.1038/bjc.2011.135 Download citation * Received: 04 January 2011 * Revised: 18 March 2011 * Accepted: 28 March 2011 * Published: 03 May 2011 * Issue Date: 24 May 2011 *
DOI: https://doi.org/10.1038/bjc.2011.135 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * HER2 * HER2/neu * ERBB2 * c-erb-b2 * heterogeneity *
real-time PCR