- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The hierarchical population structure of five, native-Spanish donkey breeds (Andaluza, Catalana, Mallorquina, Encartaciones and Zamorano-Leonesa) has been studied using
_F_-statistics. In addition, nine Moroccan asses and 24 Merens breed horses were included in the analysis. Data came from 15 DNA microsatellites. The analysis shows that Spanish donkeys are
substructured at both hierarchical levels studied, among breeds and within breeds (between subpopulations). In the whole population, the deficit of heterozygotes was estimated to be about
21%. The fixation indices corresponding to differences between breeds, subpopulations within breeds, and within subpopulations were estimated to be 6.4%, 3.5% and 3.0%, respectively. The
dendrogram obtained shows that the Andaluza and the Moroccan ass form a separate cluster from the northern Spanish breeds (Catalana, Encartaciones, Mallorquina and Zamorana-Leonesa). These
groupings coincide with those obtained from historical and archaeological data. SIMILAR CONTENT BEING VIEWED BY OTHERS AN ISOLATED POPULATION REVEALS GREATER GENETIC STRUCTURING OF THE
AUSTRALIAN DINGO Article Open access 09 November 2022 WOLF GENETIC DIVERSITY COMPARED ACROSS EUROPE USING THE YARDSTICK METHOD Article Open access 22 August 2023 MICROSATELLITE AND
MITOCHONDRIAL DNA ANALYSES UNVEIL THE GENETIC STRUCTURE OF NATIVE SHEEP BREEDS FROM THREE MAJOR AGRO-ECOLOGICAL REGIONS OF INDIA Article Open access 24 November 2020 INTRODUCTION The ass
(_Equus asinus_), is a herbivorous animal of the order _Perissodactyla_, family _Equidae_. It was domesticated about 6000 years ago, probably in either Egypt or Mesopotamia (Littauer and
Crouwel, 1979). In Spain, the development of donkey populations has been influenced by their extensive use for riding and as a beast of burden; it is capable of carrying over 100 kg. The
Spanish donkey breeds Andaluza, Catalana, Mallorquina, Encartaciones and Zamorano-Leonesa have suffered a substantial decrease in population size (Jordana and Folch, 1998) which might create
high levels of inbreeding which may result in inbreeding depression, increasing the risk of breed extinction. Currently, the census population sizes of these five breeds are very low, and
they are included in the Food and Agricultural Organization of the United Nation’s (FAO) list of domestic animals to be preserved (FAO, DAD-IS http://fao.org/dad-is). The objective of the
present study is to characterize the genetic structure of five Spanish donkey populations in danger of extinction by using _F_-statistic analysis (Wright, 1965; Nei, 1977; Weir and
Cockerham, 1984). _F_-statistics have proven to be a very useful tool in elucidating the pattern and extent of genetic variation residing within and among natural populations of animal and
plant species. Genetic characterisation is the first step in breed conservation programmes and may have implications for future breeding strategies (Bjørnstad et al, 2000). Little genetic
data from domestic donkeys exist (Breen et al, 1994; Bellone et al, 1998; Jordana et al, 1999, 2001; Aranguren-Méndez et al, 2001). MATERIALS AND METHODS BREEDS STUDIED AND DNA COLLECTION
Genomic DNA was prepared from whole blood according to standard methods involving lysates of the washed white cells and phenol-cloroform-isoamylalcohol (25:24:1) extraction. The sample size
and breeds involved were: 87 Andaluza (AND), 140 Catalana (CAT), 104 Mallorquina (MALL), 74 Encartaciones (ENC) and 108 Zamorano-Leonesa (ZAM). The areas of main distribution of these
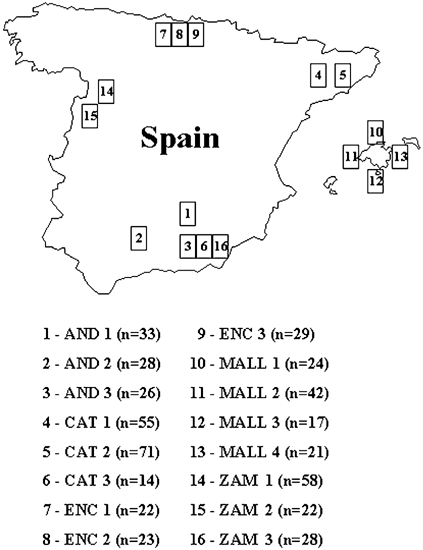
indigenous Spanish breeds are shown in Figure 1. In addition, nine Moroccan asses were used as members of the _E. asinus africanus_, and 24 horses of the Merens breed (_E. caballus_) were
used as an outgroup. The breeds were divided into 16 subpopulations, according to geographical criteria and/or areas of influence of certain breeders (Figure 1). MICROSATELLITE MARKERS The
15 equine microsatellite markers chosen for analysis were: AHT4, AHT5, ASB2, HMS1, HMS2, HMS3, HMS5, HMS6, HMS7, HTG4, HTG6, HTG7, HTG10, HTG15 and VHL20. Primer sequences, reaction
conditions and the data obtained (available from the authors on request) have been described previously by Aranguren-Méndez et al (2001). STATISTICAL ANALYSES Population structure was
analysed by means of _F_-statistics using Weir and Cockerham’s methods (1984), implemented in the FSTAT computer programme (Goudet, 2000). A hierarchical _F_ST analysis was performed to
determine the amount of variance attributable to subpopulation substructure (Wright, 1978; Johannesen and Loeschcke, 1996). The hierarchical analysis was carried out using analysis of
molecular variance (AMOVA) in the ARLEQUIN 2000 package (Schneider et al, 2000). AMOVA yields estimations of population structure at different levels of the specified hierarchy. In order to
test the significance of the different statistics for the null hypothesis of no differentiation at the corresponding level, permutation and resampling tests (jack-knifing and bootstrapping)
were carried out as implemented in the previous programmes. The Reynolds genetic distance, _D_R, (Reynolds et al, 1983), a measure based on _F_ST values [_D_R= −ln(1−_F_ST)], with
neighbour-joining (NJ) clustering (Saitou and Nei, 1987), was used to construct a dendrogram of breed relationships, using the PHYLIP package (Felsenstein, 1995). An unrooted consensus tree,
evaluated by 1000 bootstrap replicates, was obtained. The method of Slatkin (1993), implemented in GENEPOP, was used to assess the genetically effective migration rate (M = Nem, the average
number of migrants exchanged per generation). RESULTS Thirteen of 15 equine microsatellites investigated amplified well and were polymorphic in the donkey, except for locus ASB2, which
failed to amplify, and HMS1, which was monomorphic (165 bp) in all breeds. _F_ST, _F_IT and _F_IS values, computed over all breeds and loci, were obtained. Levels of apparent breed
differentiation were considerable and multilocus _F_ST values indicate that around 4.1% of the total genetic variation could be explained by breed differences, the remaining 95.9%
corresponding to differences among individuals. Genetic differentiation among breeds was highly significant (_P_ < 0.001) for all loci. On average, individuals within breeds had a 17.8%
(_P_ < 0.001) deficit of heterozygotes, whereas the total population had a 21.1% (_P_ < 0.001) deficit of heterozygotes. Values for the _F_-statistics of the Spanish donkey populations
at all hierarchical levels are presented in Table 1. The degree of genetic differentiation among subpopulations was highly significant for all breeds, varying from 1.3% for the
Encartaciones breed to 5.8% for the Andaluza breed (_P_ < 0.001). The hierarchical analysis of the Spanish donkey populations (Table 2) revealed that, as expected, most of the
differentiation occurs among breeds with respect to the total population rather than among subpopulations within breeds, and within subpopulations; 0.064 _vs_ 0.035, and 0.030, respectively.
The differentiation among breeds in the hierarchical analysis was 6.4%, more than that obtained without using a hierarchical analysis (4.1%). Table 3 shows the total inbreeding estimate
(_F_ ≅ _F_IT) per breed, evaluated by hierarchical analysis. The FIT average, obtained from jackknifing over loci, ranged from 0.112 ± 0.049 to 0.232 ± 0.058, for the Mallorquina and
Andaluza breeds, respectively. The interbreed genetic distance, or _F_ST estimates, below the diagonal, and gene flow (Nem) above the diagonal, between pairs of Spanish donkey breeds are
shown in Table 4. After 1000 permutations, performed with FSTAT, all _F_ST calculated by pairwise breed combinations were significantly different from zero (_P_ < 0.001). Least
differentiation was detected between the Andaluza and Zamorano-Leonesa breeds and Encartaciones-Mallorquina breeds (_F_ST = 0.031), and the most divergence was observed between the Andaluza
and Catalana breeds (_F_ST = 0.058). Nem represents the number of effective migrants exchanged per generation. The Nem values for pairs of breeds varied from 4.16 to 7.88 for the AND-CAT
pair and the MALL-ENC pair, respectively. Figure 2 shows a NJ tree based on Reynolds genetic distance (data not shown) relating the five Spanish donkey breeds studied, the Moroccan ass, and
the Merens breed used as outgroup. The numbers at the nodes are bootstrapping values for 1000 replicates of the 13 loci genotyped. DISCUSSION Genetic differentiation among Spanish donkey
breeds exists. All loci contribute to this differentiation with _F_ST values being moderately low and similar for all systems studied, but very significant (_P_ < 0.001). However, it is
clear that most of the total genetic variation corresponds to differences among individuals (95.9%) and only less than 5% is due to differences among breeds. These values of total genetic
differentiation (_F_ST) among breeds are close to those found in other domestic species, for example: among river buffalo breeds (_F_ST = 0.038, Barker et al, 1997), among Spanish horse
breeds (_F_ST = 0.078, Cañon et al, 2000), though they are slightly lower than those found in Norwegian horse breeds (_F_ST = 0.12, Bjørnstad et al, 2000), in European cattle breeds (_F_ST =
0.11, MacHugh et al, 1998; Kantanen et al, 2000) and among Spanish dogs (_F_ST = 0.099, Jordana et al, 1992). The results from the hierarchical analysis further show that populations of
Spanish donkeys are substructured at different levels (Table 2). The differentiation within breeds is only half of that between breeds (3.5% and 6.4%, respectively). This value (within
breeds) was similar to that reported for other organisms: Spanish dogs (Jordana et al, 1992), black-tailed dogs (Chesser, 1983) and house mice (Selander and Kaufman, 1975). The total
inbreeding estimate (_F_ ≅ _F_IT; Table 3) showed a significant deficit of heterozygotes (_P_ < 0.001) for all breeds. Consanguinity, which is produced by mating between relatives, can be
one cause of a deficit of heterozygotes, but this deficit affects all or most of the loci in a similar way. The AND, CAT and ZAM breeds showed 10 or 11 loci, of the total of 13, with a
significant deficit of heterozygotes, which might thus result from consanguinity (Table 3). Although in these breeds high and significant values of _F_ST (Table 1) (0.058, 0.031 and 0.037,
respectively), also suggest that a significant subpopulation structure (a Wahlund effect) exists. On the other hand, MALL and ENC breeds showed significant deficits of heterozygotes at only
five and six loci, respectively, these deficits being harder to explain by consanguinity. We must remember that there are other factors that can also cause a lack of heterozygotes in the
population (Nei, 1987). First, ‘null alleles’ (non-amplifying alleles) may be present and lead to a false observation of excess homozygotes. Secondly, the presence of population substructure
within the breed may lead to a Wahlund effect. Reproductive substructure within the breed is a feasible explanation for the high deficit of heterozygotes observed in the Spanish donkey
populations. Nevertheless, two loci exist, specifically HTG4 and HMS7, which show a very significant deficit of heterozygotes in all breeds (_P_ < 0.001). The most coherent interpretation
to explain this deficit is the possible presence, not detected, of ‘null alleles’ in these populations. Previous reports have indicated the occurrence of a ‘null allele’ present in the HMS7
locus in the Quarter-horse equine breed (Bozzini et al, 1997). The dendrogram’s topology (Figure 2) is very similar to that obtained by Aranguren-Mèndez et al (2001) using the _D_A genetic
distance (Nei, 1987) and the NJ algorithm. In this work, the four breeds of black coat from the north of Spain (CAT, ENC, MALL and ZAM) form a closed cluster (64% support), supporting the
hypothesis of a common ancestral past from _E. asinus europeus_. In this paper, unlike Aranguren-Mèndez et al (2001), the African origin of the Andaluza breed is clarified, which forms a
cluster with the Moroccan ass, a genuine representative of the ancestral trunk of _E. asinus africanus_. This supports the thesis earlier stated by other authors about its African origin
(Aparicio, 1960; Epstein, 1984; Sotillo and Serrano, 1985). However, the low bootstrap value for this cluster (43% support) reflects the instability of the topology, so additional studies to
confirm this hypothesis are necessary. Further investigations involving more European and African donkey populations, as well as the analysis of genetic variability of mtDNA, could look for
possible introgression of mtDNA haplotypes of African origin into these populations. REFERENCES * Aparicio, G (1960). _Zootecnia Especial_. Etnología compendiada: Imprenta Moderna Córdoba.
Google Scholar * Aranguren-Méndez, JA, Jordana, J, Gomez, M (2001). Genetic diversity in Spanish donkey breeds using microsatellite DNA markers. _Genet Sel Evol_, 33: 433–442. Article
PubMed PubMed Central Google Scholar * Barker, JSF, Moore, SS, Hetzel, DJS, Evans, S, Tan, SG, Byrne, K (1997). Genetic diversity of Asian water buffalo (_Bubalus bubalis_):
microsatellite variation and a comparison with protein-coding loci. _Anim Genet_, 28: 103–115. Article CAS PubMed Google Scholar * Bellone, RR, Cothran, EG, Ketchum, MS (1998). Genetic
variation in the rare donkey breed, Baudet du Poitou. _Anim Genet_, 29: (Suppl. 1): 17 Google Scholar * Bjørnstad, G, Gunby, E, Røed, KH (2000). Genetic structure of Norwegian horse breeds.
_J Anim Breed Genet_, 117: 307–317. Article Google Scholar * Bozzini, M, Eggleston-Stott, ML, Del Valle, A, Ziegle, JS (1997). Addition of degenerate primer in multiplex PCRs corrects the
equine microsatellite locus HMS7 null allele in the Quarter horse. Abstracts P50. Plant & Animal Genome V Conference. San Diego, CA. * Breen, M, Downs, P, Irvin, Z, Bell, K (1994).
Intrageneric amplification of horse microsatellite markers with emphasis on the Przewalski’s horse (_E. przewalskii_). _Anim Genet_, 25: 401–405. Article CAS PubMed Google Scholar *
Cañon, J, Checa, ML, Carleos, C, Vega-Pla, JL, Vallejo, M, Dunner, S (2000). The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data. _Anim Genet_, 31: 39–48.
Article PubMed Google Scholar * Chesser, RK (1983). Genetic variability within and among populations of the black-tailed prairie dog. _Evolution_, 37: 320–331. Article PubMed Google
Scholar * Epstein, H (1984). Ass, mule and onager. In: Mason IL (ed) _Evolution of Domesticated Animals_, Longman: London and New York, pp 174–184. Google Scholar * Felsenstein, J (1995).
_PHYLIP (Phylogeny inference package) version 3.57c_. Department of Genetics, University of Eashington, Seattle. Google Scholar * Goudet, J (2000). FSTAT Ver. 2.9.1. Computer package for
PCs. Institute of Ecology, Biology Building, UNIL, CH-1015 Lausanne, Switzerland. * Johannesen, J, Loeschcke, V (1996). A hierarchical analysis of genetic structure and variability in
patchily distributed coexisting _Chiastocheta_ species (Diptera: Anthomyiidae). _Heredity_, 76: 437–448. Article Google Scholar * Jordana, J, Piedrafita, J, Sanchez, A, Puig, P (1992).
Comparative F statistics analysis of the genetic structure of ten Spanish dog breeds. _J Hered_, 83: 367–374. Article CAS PubMed Google Scholar * Jordana, J, Folch, P (1998). The
Catalonian donkey breed: program of conservation and improvement of an endangered breed. _Arch Zootec_, 47: 403–409. Google Scholar * Jordana, J, Folch, P, Sanchez, A (1999). Genetic
variation (protein markers and microsatellites) in endangered Catalonian donkeys. _Bioch Syst Ecol_, 27: 791–798. Article CAS Google Scholar * Jordana, J, Folch, P, Aranguren, JA (2001).
Microsatellite analysis of genetic diversity in the Catalonian donkey breed. _J Anim Breed Genet_, 118: 57–63. Article CAS Google Scholar * Kantanen, J, Olsaker, I, Holm, LE, Lien, S,
Vilkki, J, Brusgaard, K _et al._ (2000). Genetic diversity and population structure of 20 North European cattle breeds. _J Hered_, 91: 446–457. Article CAS PubMed Google Scholar *
Littauer, MA, Crouwel, JH (1979). _Wheeled Vehicles and Ridden Animals in the Ancient Near East_. Brill: Leiden and Köln. Google Scholar * Machugh, DE, Loftus, RT, Cunningham, P, Bradley,
DG (1998). Genetic structure of seven European cattle breeds assessed using 20 microsatellite markers. _Anim Genet_, 29: 333–340. Article CAS PubMed Google Scholar * Nei, M (1977).
F-statistics and analysis of gene diversity in subdivided populations. _Ann Hum Genet_, 41: 225–233. Article CAS PubMed Google Scholar * Nei, M (1987). _Molecular Evolutionary Genetics_.
Columbia University Press: New York. Google Scholar * Reynolds, J, Weir, BS, Cockerham, CC (1983). Estimation of the coancestry coefficient: Basis for a short-term genetic distance.
_Genetics_, 105: 767–769. CAS PubMed PubMed Central Google Scholar * Saitou, N, Nei, M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. _Mol Biol
Evol_, 4: 406–425. CAS PubMed Google Scholar * Schneider, S, Roessli, D, Excoffier, L (2000). _Arlequin ver. 2000: a software for population genetics data analysis_. Genetics and Biometry
Laboratory, University of Geneva, Switzerland. Google Scholar * Selander, RB, Kaufman, DW (1975). Genetic structure of populations of the brown snail (_Helix apersa_): I. Microgeographic
variation. _Evolution_, 29: 385–401. PubMed Google Scholar * Slatkin, M (1993). Isolation by distance in equilibrium and non-equilibrium populations. _Evolution_, 47: 264–279. Article
PubMed Google Scholar * Sotillo, JL, Serrano, V (1985). Producción Animal 1, Etnología Zootécnica. Tebar-Flores: Madrid. * Weir, BS, Cockerham, CC (1984). Estimating F-statistics for the
analysis of population structure. _Evolution_, 38: 1358–1370. CAS PubMed Google Scholar * Wright, S (1965). Interpretation of population structure by F-statistics with special regard to
system of mating. _Evolution_, 19: 395–420. Article Google Scholar * Wright, S (1978). _Evolution and the genetics of populations: Vol 4 Variability within and among natural populations_.
University of Chicago Press: Chicago. Google Scholar Download references ACKNOWLEDGEMENTS This study was made possible by the financial support of the CICYT (project AGF98–0503) and the
DARP (Generalitat de Catalunya); we are also grateful to the Breed Associations of the Spanish donkeys for their helpful co-operation and assistance during the sample collection. Also, we
would like to thank Rosa Avellanet for his valuable comments on the manuscript, and Chuck Simmons for the English revision. The suggestions of the two referees were greatly appreciated.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Unitat de Genètica i Millora Animal, Departament de Ciència Animal i dels Aliments, Facultat de Veterinària, Universitat Autònoma de Barcelona,
Barcelona, Spain J Aranguren-Méndez & J Jordana * Departamento de Producción Animal, Universidad del Zulia, Facultad de Ciencias Veterinarias, Maracaibo, Venezuela J Aranguren-Méndez *
Servicio de Ganadería, Diputación Foral de Bizkaia, Avda. Lehendakari Aguirre, Bilbao, Spain M Gómez Authors * J Aranguren-Méndez View author publications You can also search for this author
inPubMed Google Scholar * M Gómez View author publications You can also search for this author inPubMed Google Scholar * J Jordana View author publications You can also search for this
author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to J Jordana. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Aranguren-Méndez, J.,
Gómez, M. & Jordana, J. Hierarchical analysis of genetic structure in Spanish donkey breeds using microsatellite markers. _Heredity_ 89, 207–211 (2002).
https://doi.org/10.1038/sj.hdy.6800117 Download citation * Received: 01 June 2001 * Accepted: 29 April 2002 * Published: 04 September 2002 * Issue Date: 01 September 2002 * DOI:
https://doi.org/10.1038/sj.hdy.6800117 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * donkey * extinction danger * population structure *
_F_-statistics * hiearchical analysis * microsatellite