- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
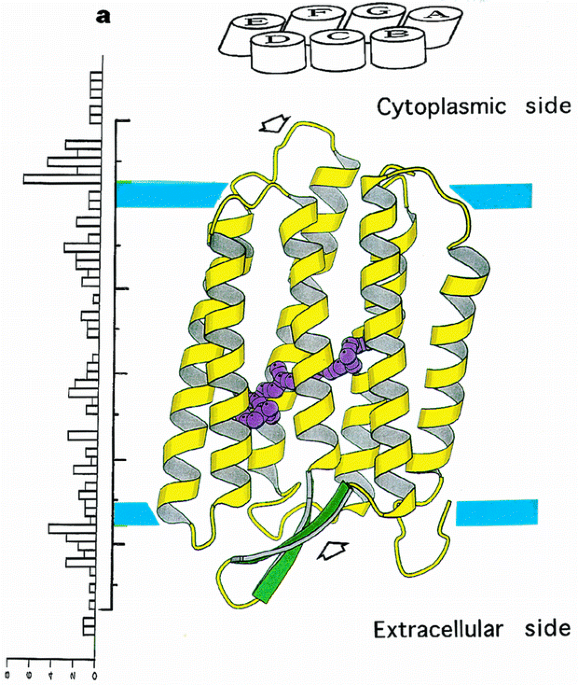
ABSTRACT Bacteriorhodopsin is a transmembrane protein that uses light energy, absorbed by its chromophore retinal, to pump protons from the cytoplasm of bacteria such as _Halobacterium
salinarium_ into the extracellular space1,2. It is made up of seven α-helices, and in the bacterium forms natural, two-dimensional crystals called purple membranes. We have analysed these
crystals by electron cryo-microscopy to obtain images of bacteriorhodopsin at 3.0 å resolution. The structure covers nearly all 248 amino acids, including loops outside the membrane, and
reveals the distribution of charged residues on both sides of the membrane surface. In addition, analysis of the electron-potential map produced by this method allows the determination of
the charge status of these residues. On the extracellular side, four glutamate residues surround the entrance to the proton channel, whereas on the cytoplasmic side, four aspartic acids
occur in a plane at the boundary of the hydrophobic–hydrophilic interface. The negative charges produced by these aspartate residues is encircled by areas of positive charge that may
facilitate accumulation and lateral movement of protons on this surface. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS STRUCTURAL BASIS FOR THE PROLONGED PHOTOCYCLE OF SENSORY RHODOPSIN
II REVEALED BY SERIAL SYNCHROTRON CRYSTALLOGRAPHY Article Open access 11 April 2025 MECHANISMS OF INWARD TRANSMEMBRANE PROTON TRANSLOCATION Article 29 June 2023 CRYO-EM STRUCTURE AND
DYNAMICS OF THE GREEN-LIGHT ABSORBING PROTEORHODOPSIN Article Open access 05 July 2021 REFERENCES * Oesterhelt, D. & Stoeckenius, W. Rhodopsin-like protein from the purple membrane of
_Halobacterium halobium_. _Nature New Biol._ 233, 149–152 (1971). Article CAS Google Scholar * Oesterhelt, D. & Stoeckenius, W. Functions of a new photoreceptor membrane. _Proc. Natl
Acad. Sci. USA_ 70, 2853–2857 (1973). Article ADS CAS Google Scholar * Khorana, H. G. Bacteriorhodopsin, a membrane protein that uses light to translocate protons. _J. Biol. Chem._ 263,
7439–7442 (1988). CAS PubMed Google Scholar * Lanyi, J. K. Proton translocation mechanism and energetics in the light-driven pump bacteriorhodopsin. _Biochim. Biophys. Acta_ 1183, 241–261
(1993). Article CAS Google Scholar * Henderson, R. _et al_. Amodel for the structure of bacteriorhodopsin based on high resolution electron cryo-microscopy. _J. Mol. Biol._ 213, 899–929
(1990). Article CAS Google Scholar * Grigorieff, N., Ceska, T. A., Downing, K. H., Baldwin, J. M. & Henderson, R. Electron-crystallographic refinement of the structure of
bacteriorhodopsin. _J. Mol. Biol._ 259, 393–421 (1996). Article CAS Google Scholar * Fujiyoshi, Y. _et al_. Development of a superfluid helium stage for high-resolution electron
microscopy. _Ultramicroscopy_ 38, 241–251 (1991). Article Google Scholar * Sakata, K., Tahara, Y., Morikawa, K., Fujiyoshi, Y. & Kimura, Y. Amethod for observing cross-sectional views
of biomembranes. _Ultramicroscopy_ 45, 253–261 (1992). Article CAS Google Scholar * Gerwert, K., Hess, B., Soppa, J. & Oesterhelt, D. The role of 96 Asp in proton translocation by
bacteriorhodopsin. _Proc. Natl Acad. Sci. USA_ 86, 4943–4947 (1989). Article ADS CAS Google Scholar * Otto, H. _et al_. Aspartic acid-96 is the internal proton donor in the reprotonation
of the Schiff base of bacteriorhodopsin. _Proc. Natl Acad. Sci. USA_ 86, 9228–9232 (1989). Article ADS CAS Google Scholar * Braiman, M. S. _et al_. Vibrational spectroscopy of
bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. _Biochemistry_ 27, 8516–8520 (1988). Article CAS Google
Scholar * Kimura, Y. & Ikegami, A. Local dielectric properties around polar region of lipid bilayer membranes. _J. Membr. Biol._ 85, 225–231 (1985). Article CAS Google Scholar *
Brown, L. S. _et al_. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. _J. Biol. Chem._ 270, 27122–27126 (1995). Article CAS Google
Scholar * Kushwaha, S. C., Kates, M. & Stoeckenius, W. Comparison of purple membrane from _Halobacterium cutirubrum_ and _Halobacterium halobium_. _Biochim. Biophys. Acta_ 426, 703–710
(1976). Article CAS Google Scholar * Subramaniam, S., Greenhalgh, D. A. & Khorana, H. G. Aspartic acid 85 in bacteriorhodopsin functions both as proton acceptor and negative
counterion to the Schiff base. _J. Biol. Chem._ 267, 25730–25733 (1992). CAS PubMed Google Scholar * Riesle, J., Oesterhelt, D., Dencher, N. A. & Heberle, J. D38 is an essential part
of the proton translocation pathway in bacteriorhodopsin. _Biochemistry_ 35, 6635–6643 (1996). Article CAS Google Scholar * Mogi, T., Stern, L. J., Marti, T., Chao, B. H. & Khorana,
H. G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. _Proc. Natl Acad. Sci. USA_ 85, 4148–4152 (1988). Article ADS CAS Google Scholar * Oesterhelt, D.
& Stoeckenius, W. Isolation of the cell membrane of _Halobacterium halobium_ and its fractionation into red and purple membrane. _Methods Enzymol._ 31, 667–678 (1974). Article CAS
Google Scholar * Seiff, F., Wallat, I., Ermann, P. & Heyn, M. Aneutron diffraction study on the location of the polyene chain of retinal in bacteriorhodopsin. _Proc. Natl Acad. Sci.
USA_ 82, 3227–3231 (1985). Article ADS CAS Google Scholar * Baldwin, J. & Henderson, R. Measurement and evaluation of electron diffraction patterns from two-dimensional crystals.
_Ultramicroscopy_ 14, 319–333 (1984). Article CAS Google Scholar * Ceska, T. A. & Henderson, R. Analysis of high-resolution electron diffraction patterns from purple membrane labeled
with heavy atoms _J. Mol. Biol._ 213, 539–560 (1990). Article CAS Google Scholar * Collaborate Computational Project No. 4 _Acta Crystallogr. D_ 50, 760–763 (1994). * International Union
of Crystallography _International table for Crystallography Volume C: Mathematical, Physical and Chemical Table_ (corrected edn) (ed. Wilson, A. J. C.) (Kluwer, Dordrecht, 1995). Google
Scholar Download references ACKNOWLEDGEMENTS We thank M. Ikehara for encouragement and support; W. Kühlbrandt and D. N. Wang for help with data processing; R. Henderson, S. Fuller, J.
Lanyi, W. Stoeckenius, Y. Harada and J.Sasaki for helpful discussions; T. Miyata for preparing this manuscript; and Digital Equipment Corporation for help with computers. AUTHOR INFORMATION
Author notes * Yoshinori Fujiyoshi Present address: Department of Biophysics, Faculty of Science, Kyoto University, Oiwake Kitashirakawa, Sakyou-ku, 606-01, Kyoto, Japan * Dmitry G.
Vassylyev Present address: International Institute for Advanced Research (IIAR), Matsushita Electric Industrial Co., Ltd., 3-4 Hikaridai, Seika, Soraku, Kyoto, 619-02, Japan * Atsuo Miyazawa
Present address: MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 2QH, UK * Akinori Kidera Present address: Department of Chemistry, Faculty of Science, Kyoto University,
Oiwake Kitashirakawa, Sakyou-ku, Kyoto, 606-01, Japan AUTHORS AND AFFILIATIONS * Biomolecular Engineering Research Institute (formerly Protein Engineering Institute), 6-2-3 Fureudai, Suita,
565, Osaka, Japan Yoshiaki Kimura, Dmitry G. Vassylyev, Atsuo Miyazawa & Akinori Kidera * Rational Drug Design Laboratory, 4-1-4 Misato, Matsukawa, 960-12, Fukushima, Japan Masaaki
Matsushima * International Institute for Advanced Research, Matsushita Electric Industrial Co., 3-4 Hikaridai, Seika, Soraku, 619-02, Kyoto, Japan Kaoru Mitsuoka, Kazuyoshi Murata, Teruhisa
Hirai & Yoshinori Fujiyoshi * Department of Biophysics, Faculty of Science, Kyoto University, Oiwake Kitashirakawa, Sakyou-ku, 606-01, Kyoto, Japan Dmitry G. Vassylyev, Atsuo Miyazawa
& Akinori Kidera Authors * Yoshiaki Kimura View author publications You can also search for this author inPubMed Google Scholar * Dmitry G. Vassylyev View author publications You can
also search for this author inPubMed Google Scholar * Atsuo Miyazawa View author publications You can also search for this author inPubMed Google Scholar * Akinori Kidera View author
publications You can also search for this author inPubMed Google Scholar * Masaaki Matsushima View author publications You can also search for this author inPubMed Google Scholar * Kaoru
Mitsuoka View author publications You can also search for this author inPubMed Google Scholar * Kazuyoshi Murata View author publications You can also search for this author inPubMed Google
Scholar * Teruhisa Hirai View author publications You can also search for this author inPubMed Google Scholar * Yoshinori Fujiyoshi View author publications You can also search for this
author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Yoshiaki Kimura. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kimura, Y.,
Vassylyev, D., Miyazawa, A. _et al._ Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. _Nature_ 389, 206–211 (1997). https://doi.org/10.1038/38323 Download
citation * Received: 21 October 1996 * Accepted: 02 June 1997 * Issue Date: 11 September 1997 * DOI: https://doi.org/10.1038/38323 SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative