- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
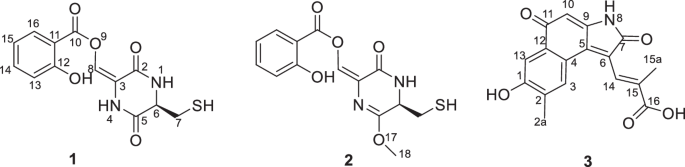
ABSTRACT Three new compounds including two thiodiketopiperazine derivatives shovelmycins A–B (1–2), and one ansamycin derivative divergolide X (3) were isolated and identified from the
culture extract of _Streptomyces olivaceus_ HDN22-001, a marine actinomycete obtained from the deep-sea cold seep sediment sample collected from the South China Sea. Their structures and
absolute configurations were determined by spectroscopic analyses and ECD calculations. Compound 1 exhibited the strongest DPPH radical scavenging activity with an IC50 value of 10.83 μM,
which was better than that of the positive control vitamin C. And compound 2 was modestly cytotoxic against NCl-H446 cell with the IC50 value of 26.6 μM. You have full access to this article
via your institution. Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS A NEW STEROID WITH POTENT ANTIMICROBIAL ACTIVITIES AND TWO NEW POLYKETIDES FROM _PENICILLIUM VARIABILE_ EN-394, A
FUNGUS OBTAINED FROM THE MARINE RED ALGA _RHODOMELA CONFERVOIDES_ Article 26 October 2023 ANTIBACTERIAL _P_-TERPHENYL AND _Α_‑PYRONE DERIVATES ISOLATED FROM THE MARINE-DERIVED ACTINOMYCETE
_NOCARDIOPSIS_ SP. HDN154086 Article 25 January 2024 RAUSUQUINONE, A NON-GLYCOSYLATED PLURAMYCIN-CLASS ANTIBIOTIC FROM _RHODOCOCCUS_ Article 30 November 2021 INTRODUCTION Deep-sea cold seeps
represented extreme environments characterized by the expulsion of hydrocarbon-rich fluids, hydrogen sulfide, and fine-grained sediments [1,2,3,4,5]. These distinct ecosystems provided
abundant carbon and sulfur sources that support diverse microbial communities, leading to the production of novel secondary metabolites with significant biological activities, including
antimicrobial, cytotoxic, neuraminidase inhibitory, and antioxidant properties [6,7,8,9]. Sulfur-containing metabolites with potent biological activities were frequently encountered in
marine microbial natural products isolated from cold seep environments. Notable examples include verruculosa A, isolated from _Curvularia verruculosa_ CS-129, which exhibits significant
inhibitory activity against pathogenic _Escherichia coli_ [10]. Similarly, pseudallenes A and B, novel compounds isolated from _Pseudallescheria boydii_ CS-793, demonstrate broad-spectrum
antimicrobial activity against various phytopathogens [11]. Despite these findings, sulfur-containing compounds from cold seep-associated actinomycetes were considerably underrepresented
compared to their fungal counterparts, only one case has been reported [12]. In addition, studies examining natural products from cold seep actinomycetes remain notably limited, with only
two publications documenting a total of twelve compounds, including six novel structures [12, 13]. Therefore, we focused on cold seeps derived actinomycetes to explore unique structures.
RESULTS AND DISCUSSION Our investigation with a cold seep sediment strain, HDN22-001, identified as a _Streptomyces olivaceus_, was found to produce two new sulfur-containing compounds,
shovelmycins A and B (1–2), a new opened linear ansamycin analog divergolide X (3). In this study, the fermentation, isolation, structural elucidation and bioactivity of these compounds
(Fig. 1) were described. Compound 1 was separated as a yellow amorphous powder, with the chemical formula C13H12N2O5S based on HRESIMS spectrum (307.0388, [M − H]−), indicating nine degrees
of unsaturation. The IR absorptions at 3672 and 1681 cm–1 indicated the presence of hydroxy and carbonyl groups, respectively. In the 1H NMR spectrum collected in DMSO-_d_6, two broad
singlets at _δ_H 13.05 (12-OH), 10.41 (4-NH), and one broad doublet at _δ_H 8.98 (1-NH) were ascribable to three exchangeable protons. 13C NMR data displayed 13 carbon resonances (Table 1),
combined with the HSQC cross-peaks, accounting for 1 methylene, 6 methines (with 5 aromatic/olefinic), and 6 nonprotonated carbons (with 3 aromatic/olefinic and 3 carbonyls). Detailed
analysis of the 1D and 2D NMR data allowed the establishment of fragments 1 A and 1B for compound 1 (Fig. 2). Fragment 1 A was deduced as salicylic acid similar fragment [14] based on the 1D
NMR spectrum data. Fragment 1B was deduced to be a diketopiperazine derivative similar to cyclo(l-Cys-l-Leu) [15], except that the signals of isobutyl group carbons in the 13C NMR spectrum
of cyclo(l-Cys-l-Leu) were replaced by two olefinic carbons, with one nonprotonated carbon (_δ_C 136.9, C-3 in methanol-_d_4) and one methine (_δ_C 142.4, CH-8, in methanol-_d_4),
respectively. Also, the key HMBC correlation from H-8 (_δ_H 8.54) to C-2 (_δ_C 162.4) and C-3 (_δ_C 136.9) further determined the assignment of fragment 1B. The conjunction between 1 A and
1B was through ester bond of C-8 − O − C-10, supported by the key HMBC correlation from H-8 to C-10 (_δ_C 162.6). Thus, the planar structure of 1 was constructed (Fig. 3). Due to the free
rotation of the C-6 (_δ_C 56.2) and C-7 (_δ_C 26.4) single bond and the _cis-trans_ isomerization of the C-3 and C-8 (_δ_C 142.4) double bond, it was a challenge to determine the absolute
configurations of 1. There were four relative configurations, named (3_Z_*, 6_R_*)-1A, (3_E_*, 6_R_*)-1B, (3_E_*, 6_S_*)-1C and (3_Z_*, 6_S_*)-1D, theoretically. To determine the _cis-trans_
isomerization of the C-3 and C-8 double bond, the 13C NMR chemical shifts for the four possible isomers were calculated at the B3LYP/6-31 + G (d)//B3LYP/6-311 + G (d, p) levels and further
checked by DP4+ probability. The (3_E_*, 6_R_*)-1 isomer showed a striking predominance (89.83% probability) over the other three configurations. In addition, the total probability of the
3_E_* configurations reach an astonishing 94.56%, far exceeding the probability of the 3_Z_* configuration, which allowed us to assign the relative configuration of 1 as 3_E_*. To determine
the absolute configuration of C-3 and C-6 in 1, the ECD calculations of the optimized conformation of (3_E_, 6_R_)-1 obtained at the B3LYP/6-31 + G(d) level was performed. The overall
pattern of the experimental ECD spectrum was in reasonable agreement with the calculated one of (3_E_, 6_R_)-1 (Fig. 4), indicating the absolute configuration of 3_E_, 6_R_ in 1. Compound 2
was separated as a yellow amorphous powder, with the chemical formula C14H14N2O5S based on HRESIMS spectrum (321.0546, [M − H]−), with nine degrees of unsaturation. The 1D NMR data of 2
(Table 1) were highly similar to the co-isolated 1. The main distinctions between 2 and 1 were that the carbonyl group of the C-5 (_δ_C 172.9) in 1 become the hydroxyl group due to the
tautomerism of keto-enol, and then 5-OH was methylated, which can be determined by the HMBC signals from H-18 (_δ_H 3.80) to C-5 (_δ_C 171.9) and the 1D NMR data of 5-OCH3 (_δ_C 53.1) in 2
(Fig. 3). The absolute configuration of the C-3 and C-6 was determined to be 3_E_, 6 _R_ by similar experimental ECD fitting curves of compounds 2 and 1 (Figure S4). Compound 3 was separated
as a red amorphous powder, with the chemical formula C17H13NO5 based on HRESIMS spectrum (310.0723, [M − H]−), with twelve degrees of unsaturation. In the 1H NMR spectrum collected in
DMSO-_d_6, three singlets at _δ_H 10.53 (1-OH), _δ_H 10.82 (8-NH), and _δ_H 12.92 (16-COOH) were ascribable to three exchangeable protons. The 1D NMR data of 3 (Table 1) were similar to
those of divergolide R skeleton [16]. The difference was that the side chain of divergolide R was shortened and substituted by an acrylic acid group, which was supported by the molecular
formula (C17H13NO5) given by HRESIMS data, the chemical shift of C-16 (_δ_C 168.0), and an obvious carboxylic acid exchangeable proton signal appear at _δ_H 12.92 in 1H NMR spectrum.
Compounds 1–3 were further evaluated for their 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity. The results (Table 2) showed that compound 1 had intense DPPH radical
scavenging activities, with IC50 values 10.83 μM, which was significantly better than that of the positive control vitamin C (IC50 = 12.30 μM), and DPPH scavenging rate reached 89.00%.
Compound 2 had strong DPPH radical scavenging activities, with IC50 values 32.50 μM, and DPPH scavenging rate reached 83.20%. The potency of compound 1 should due to the thiol group on the
side chain to form S–S linkage to exhibit the radical scavenging effect [17]. Compounds 1−3 were tested for antimicrobial activity on seven pathogenic microorganisms (_Acinetobacter
baumannii, Klebsiella Pneumoniae, Salmonella enteritidis, Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus_ and _Enterococcus faecalis_), while none of them were obviously
active at the concentration of 64 μg mL-1. The cytotoxic activity was also evaluated against five cancer cell lines, including K562, NCl-H446/EP, L-02, NCl-H446, and MDA-MB-231. As a result
(Table S1), compound 2 had a certain activity to NCl-H446 with IC50 value of 26.6 μM, while 1 and 3 had no active (IC50 > 30.0 μM). In summary, three new compounds (1–3) were obtained and
characterized from the cultures of the cold-seep derived actinomycete _Streptomyces olivaceus_ HDN22-001. The skeleton of 1–2 was defined to possess a rare scaffold, composed of two parts,
sulfur-containing diketopiperazine and salicylic acid, naturally occurring for the first time. Compound 3 was the shortest chain length of open-chain divergolide compounds discovered so far,
and might be the precursor of biosynthesis of this class of compounds. Compound 2 showed promising antioxidant activity with IC50 value 32.50 μM, and 1 exhibited notable antioxidant
activity with IC50 value 10.83 μM, which surpassed the positive control vitamin C. To the best of my knowledge, there had been only one report of sulfur-containing compound in actinomycete
derived from cold seeps. In our study, two novel sulfur-containing active compounds were isolated, which highlighted the potential to screen and develop useful molecules from
cold-seep-derived actinomycetes. EXPERIMENTAL GENERAL EXPERIMENTAL PROCEDURES HRESIMS data were detected on a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
By using Acquity UPLC H-Class coupled to a SQ Detector 2 mass spectrometer (Waters, BEH C18 column, 1.7 μm, 2.1 × 50 mm), we recorded the LC-MS data in ESI mode. UV spectra were performed
using Waters 2487 from 200 to 600 nm (Waters Corporation, Milford, MA, USA). The optical rotations and ECD spectra were carried out on a JASCO P-1020 digital polarimeter and a JASCO J-815
spectropolarimeter, respectively, both developed by JASCO Corporation, Tokyo, Japan. Column chromatography (CC) was performed with silica gel (100-200 mesh, 200–300 mesh, Qingdao Marine
Chemical Industrials) and Sephadex LH-20 (Amersham Biosciences, San Francisco, CA, USA). 1D NMR and 2D NMR spectra were obtained on a Bruker AVANCE NEO 400 MHz spectrometer (Bruker
Corporation, Karlsruhe, Germany) and a Bruker AVANCE NEO 500 MHz spectrometer (Bruker Corporation, Karlsruhe, Germany), both using TMS as an internal standard. IR spectra were taken on a
Bruker Tensor-27 spectrophotometer (Bruker Corporation, Karlsruhe, Germany) in KBr discs. ACTINOMYCETE MATERIAL AND FERMENTATION _Streptomyces olivaceus_ HDN22-001 (GenBank accession No.
PQ517226) was isolated from a sediment sample collected from a cold-seep mud sample from the South China Sea (22°06′55.944″N,119°17′7.8″E). The strain was deposited at the Key Laboratory of
Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, People’s Republic of China. FERMENTATION The spores of _Streptomyces
olivaceus_ HDN22-001 were inoculated into 500-mL Erlenmeyer flasks containing 100 mL of culture medium comprised of 2% soluble starch, 0.1% saltpeter, 0.5% peptone, 0.05% dipotassium
phos-phate, 0.05% magnesium sulfate, 0.05% sodium chloride, and 0.001% ferrous sulfate, pH = 7.4 (in seawater collected from Huiquan Bay, Yellow Sea), and cultured at 28 °C for 6 days on a
rotary shaker at 180 rpm. A total of 40 L of broth was extracted with EtOAc (3 × 20 L) to generate the extract (15 g). EXTRACTION AND PURIFICATION For visualization, isolation and
purification, the extract was applied to a VLC (vacuum liquid chromatography) column using a stepped gradient elution of MeOH - H2O, yielding eight subfractions (Fr.1–Fr.10). Fr.5 was
separated by LH-20 column (MeOH) to obtain three sub-fractions (Fr.3.1–Fr.3.5). Fr.3.2 was purified by semi-preparative HPLC with MeCN-H2O (v/v) (45:55) to afford 1 (5.2 mg, _t_R = 16.2 min)
and 2 (2.0 mg, _t_R = 23.8 min). Fr.3.3 was purified by semi-preparative HPLC with MeCN-H2O (v/v) (35:65) to afford 3 (5.2 mg, _t_R = 15.9 min). SHOVELMYCIN A (1) Yellow powder; [_α_]25D
–17.2 (_c_ 0.1, MeOH); UV (MeOH) _λ_max (log _ε_) 216 (0.95), 262 (1.00), 307(0.69) nm; ECD (_c_ 0.5 mM, MeOH) _λ_max (Δ_ε_) 228(-3.12), 244(-2.90), 263(-3.67), 290 (-1.25), 308 (-1.58), 352
( + 0.21)nm; IR (KBr) _ν_max 3272, 1681, 1628, 1601, 1517, 1488, 1403, 1384, 1209, 1142, 953, 725 cm-1; 1H and 13C NMR data, see Table 1; HRESIMS [M – H]– _m/z_ 307.0388 (calcd for
C13H11O5N2S, 307.0394). SHOVELMYCIN B (2) Yellow powder; [_α_]25D –23.6 (_c_ 0.1, MeOH); UV (MeOH) _λ_max (log _ε_) 217(0.82), 262(0.95), 301(0.60); ECD (_c_ 0.5 mM, MeOH) _λ_max (Δ_ε_)
229(-2.90), 249(-2.12), 272(-3.50), 289 (-2.03), 305(-1.90), 352 ( + 0.54) nm; IR (KBr) _ν_max 3379, 1682, 1520, 1443, 1384, 1209, 1143, 952, 845, 801, 725 cm–1; 1H and 13C NMR data, see
Table 1; HRESIMS [M – H]– _m/z_ 321.0546 (calcd for C14H13O5N2S, 321.0545). DIVERGOLIDE X (3) Red powder; [_α_]25D –2.3 (_c_ 0.1, MeOH); UV (MeOH) _λ_max (log _ε_) 333 (0.80) nm; IR (KBr)
_ν_max 3420, 1700, 1599, 1577, 1384, 1206, 1139 cm-1; 1H and 13C NMR data, see Table 2; HRESIMS [M – H]– _m/z_ 310.0723 (calcd for C17H12O5N, 310.0715). COMPUTATION SECTION Conformational
searches were run, employing Spartan’14, based on the MMFF (Merck Molecular Force Field). All conformers were further optimized with DFT calculations at the B3LYP/6-31 + G(d) level by using
the Gaussian 09 program [18]. TDDFT calculations were performed on the fourteen lowest-energy conformations for 1 ( > 5% population). ECD spectra were obtained on the program SpecDis [19]
by using a Gaussian band shape with a 0.4 eV width for 1 from dipole-length rotational strengths. The calculated spectra were shifted by 25 nm for 1 to facilitate comparison to the
experimental data. DPPH SCAVENGING ACTIVITIES The free radical scavenging activity was examined via DPPH (2, 2-diphenyl-1-picrylhydrazyl) assay in order to determine the antioxidant
efficiency of compounds 1–3. Based on the method of Sharma [20], with some amendments, the eliminating activity against DPPH radicals was enforced. The compounds 1–3 and vitamin C were
dissolved in absolute ethanol and diluted into 7 gradients. These were vortexed to mix well, protected from light at room temperature for 30 min, and the absorbance was read at 515 nm as a
positive control. IC50 value, defined as the amount of antioxidant necessary to decrease the initial DPPH concentration by 50%. This value is calculated by plotting inhibition percentage
against extract concentration. The ability to scavenge the DPPH was calculated according to the equation: DPPH free radical scavenging rate D VC% = [(Ablank − Acontrol) ÷ Ablank] × 100% DPPH
free radical scavenging rate D sample%= [[Ablank − (Asample − Acontrol)] ÷ Ablank] × 100% Acontrol: the absorbance of the DPPH solution; Ablank: the absorbance of ethanol. ANTIMICROBIAL AND
CYTOTOXIC ACTIVITY This antimicrobial activity experiment evaluated the antibacterial activity of compounds 1−3 against six pathogenic bacteria, including gram-positive and gram-negative
bacteria such as _Acinetobacter baumannii, Klebsiella Pneumoniae, Salmonella enteritidis, Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus_ and _Enterococcus faecalis_. All
these strains were taken from the Marine Medicinal Biological Resources Center, 5# Yushan Road, Ocean University of China, Qingdao, China. The test was conducted using a conventional broth
dilution assay and ciprofloxacin was used as a positive control. The antibacterial activity of these compounds was determined by measuring the minimum inhibitory concentration (MIC). The
detailed methodologies for biological testing have been described in previous report [21]. Cytotoxicity activity of 1−3 was evaluated against K562 cell by the MTT method; NCl-H446/EP, L-02,
NCl-H446, and MDA-MB-231 cells by the SRB method. Adriamycin was used as a positive control. The detailed methodologies for biological testing have been described in a previous report [22,
23]. REFERENCES * Cong M, Pang X, Zhao K, Song Y, Liu Y, Wang J. Deep-sea natural products from extreme environments: cold seeps and hydrothermal vents. Mar Drugs. 2022;20:404. CAS PubMed
PubMed Central Google Scholar * Rampelotto PH. Extremophiles and extreme environments. Life. 2013;3:482–5. PubMed PubMed Central Google Scholar * Brown PD, Lawrence AL. The importance
of asking “how and why?” in natural product structure elucidation. Nat Prod Rep. 2017;34:1193–202. CAS PubMed Google Scholar * Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine
animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–40. CAS PubMed Google Scholar * Levin LA. Ecology of cold seep sediments: interactions of fauna with flow,
chemistry and microbes. Oceanogr Mar Biol. 2005;43:1–46. Google Scholar * Chi LP, Li XM, Wan YP, Li X, Wang BG. Ophiobolin sesterterpenoids and farnesylated phthalide derivatives from the
deep sea cold-seep-derived fungus _Aspergillus insuetus_ SD-512. J Nat Prod. 2020;83:3652–60. CAS PubMed Google Scholar * Choi BK, Trinh PTH, Lee HS, Choi BW, Kang JS, Ngoc NTD, et al.
New ophiobolin derivatives from the marine fungus _Aspergillus flocculosus_ and their cytotoxicities against cancer cells. Mar Drugs. 2019;17:346. CAS PubMed PubMed Central Google Scholar
* Li X, Li L, Li XM, Li HL, Konuklugil B, Wang BG. Ustusaustin A: a new neuraminidase inhibitory meroterpene from the ascidian-derived endophytic fungus _Aspergillus ustus_ TK-5. Nat Prod
Res. 2021;35:4939–44. CAS PubMed Google Scholar * Che YH, Wang JF, Ding WP, Xiao ZH, Shi XF, Wu JM, et al. New diketopiperazine alkaloids from the Haima cold seep-derived fungus
_Toxicocladosporium_ sp. CYH-18. Phytochem Lett. 2024;60:96–100. CAS Google Scholar * Hu XY, Wang CY, Li XM, Yang SQ, Li X, Wang BG, et al. Cytochalasin derivatives from the endozoic
_Curvularia verruculosa_ CS-129, a fungus isolated from the deep-sea squat lobster _Shinkaia crosnieri_ living in the cold seep environment. J Nat Prod. 2021;84:3122–30. CAS PubMed Google
Scholar * Ying Z, Li XM, Yang SQ, Li HL, Li X, Wang BG, et al. Pseudallenes A and B, new sulfur-containing ovalicin sesquiterpenoid derivatives with antimicrobial activity from the deep-sea
cold seep sediment-derived fungus _Pseudallescheria boydii_ CS-793. Beilstein J Org Chem. 2024;20:470–8. CAS PubMed PubMed Central Google Scholar * Dong Y, Wang JingE, Bao R, Li Y. H.
New Olimycins from a Cold‐Seep‐Derived _Streptomyces olivaceus_. Chem Biodivers. 2023;20:e202300689. CAS PubMed Google Scholar * Jin E, Li H, Liu Z, Xiao F. Antibiotic Dixiamycins from a
Cold-Seep-Derived _Streptomyces olivaceus_. J Nat Prod. 2021;84:2606–11. CAS PubMed Google Scholar * Chen CL, Hu SJ, Zhu LL, Yao LM, Wang XX, Zhang AN, et al. Isolation and identification
of chemical constituents of _Rubi Fructus_, screening novel cannabinoid CB2 receptor agonist and evaluation of its anti-osteoporosis effect. Chin Tradit Herbal Drugs. 2024;55:386–401.
Google Scholar * Furukawa T, Akutagawa T, Funatani H, Uchida T, Hotta Y, Niwa M, et al. Cyclic dipeptides exhibit potency for scavenging radicals. Bioorg Med Chem. 2012;20:2002–9. CAS
PubMed Google Scholar * Zhao G, Li S, Guo Z, Sun M, Lu C. Overexpression of div 8 increases the production and diversity of divergolides in _Streptomyces_ sp. W112. RSC adv.
2015;5:98209–14. CAS Google Scholar * Tu L, Shen S, Yan Z, Li X, Liu K, Xu J, et al. Discovery of olimycin E from _Streptomyces_ sp. 11695. Nat Prod Res. 2024;4:1–6. Google Scholar *
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09, Revision A.1, Gaussian Inc. Wallingford CT;USA;2009. * Bruhn T, Schaumlöffel A, Hemberger Y,
Bringmann G SpecDis, Version 1.53. University of Wuerzburg;Germany;2011. * Sharma OP, Tej KB. DPPH antioxidant assay revisited. Food Chem. 2009;113:1202–5. CAS Google Scholar * Andrews JM.
Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. CAS PubMed Google Scholar * Mosmann T. Rapid colorimetric assay for cellular growth and
survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. CAS PubMed Google Scholar * Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica
D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This work
was supported by the Fundamental Research Funds for the Central Universities (202362008, 202172002), the Shandong Provincial Natural Science Foundation (ZR2021MH257), the National Key
Research and Development Program of China (2022YFC2804400). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of Medicine and Pharmacy, Key Laboratory of Marine Drugs Ministry of
Education, Frontiers Science Center for Deep Ocean Multispheres and Earth System, Ocean University of China, Qingdao, People’s Republic of China Chen Li, Jiangli Cheng, Ruojin Liu, Xiaofei
Huang, Luning Zhou, Guojian Zhang, Tianjiao Zhu, Dehai Li & Qian Che * Laboratory for Marine Drugs and Bioproducts, Laoshan Laboratory, Qingdao, People’s Republic of China Guojian Zhang,
Tianjiao Zhu, Dehai Li & Qian Che Authors * Chen Li View author publications You can also search for this author inPubMed Google Scholar * Jiangli Cheng View author publications You can
also search for this author inPubMed Google Scholar * Ruojin Liu View author publications You can also search for this author inPubMed Google Scholar * Xiaofei Huang View author
publications You can also search for this author inPubMed Google Scholar * Luning Zhou View author publications You can also search for this author inPubMed Google Scholar * Guojian Zhang
View author publications You can also search for this author inPubMed Google Scholar * Tianjiao Zhu View author publications You can also search for this author inPubMed Google Scholar *
Dehai Li View author publications You can also search for this author inPubMed Google Scholar * Qian Che View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to Qian Che. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION TABLE S1 RIGHTS AND PERMISSIONS Springer Nature or its
licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the
accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li,
C., Cheng, J., Liu, R. _et al._ The thiodiketopiperazine derivatives shovelmycin A–B and ansamycin derivative divergolide X from the cold-seep-derived _Streptomyces olivaceus_ HDN22-001. _J
Antibiot_ 78, 275–280 (2025). https://doi.org/10.1038/s41429-025-00812-z Download citation * Received: 28 November 2024 * Revised: 23 January 2025 * Accepted: 16 February 2025 * Published:
04 March 2025 * Issue Date: April 2025 * DOI: https://doi.org/10.1038/s41429-025-00812-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative











